Juicer Gene Expression
Ittai Eres
2018-09-14
Last updated: 2018-09-28
Code version: e0ca9da
Preparing the gene expression data
Here I read in a dataframe of counts summarized at the gene-level, then do some pre-processing and normalization to obtain a voom object to later run linear modeling analyses on. This is identical code to what was done in the case with HOMER, since this gene expression analysis is completely orthogonal to the Hi-C normalization and significance calling schemes employed.
#Read in counts data, create DGEList object out of them and convert to log counts per million (CPM). Also read in metadata.
counts <- fread("~/Desktop/Hi-C/gene_expression/counts_iPSC.txt", header=TRUE, data.table=FALSE, stringsAsFactors = FALSE, na.strings=c("NA",""))
colnames(counts) <- c("genes", "C-3649", "G-3624", "H-3651", "D-40300", "F-28834", "B-28126", "E-28815", "A-21792")
rownames(counts) <- counts$genes
counts <- counts[,-1]
dge <- DGEList(counts, genes=rownames(counts))
#Now, convert counts into RPKM to account for gene length differences between species. First load in and re-organize metadata, then the gene lengths for both species, and then the function to convert counts to RPKM.
meta_data <- fread("~/Desktop/Hi-C/gene_expression/Meta_data.txt", sep="\t",stringsAsFactors = FALSE,header=T,na.strings=c("NA",""))
meta_data$fullID <- c("C-3649", "H-3651", "B-28126", "D-40300", "G-3624", "A-21792", "E-28815", "F-28834")
ord <- data.frame(fullID=colnames(counts)) #Pull order of samples from expression object
left_join(ord, meta_data, by="fullID") -> group_ref #left join meta data to this to make sure sample IDs correctWarning: Column `fullID` joining factor and character vector, coercing into
character vector#Read in human and chimp gene lengths for the RPKM function:
human_lengths<- fread("~/Desktop/Hi-C/gene_expression/human_lengths.txt", sep="\t",stringsAsFactors = FALSE,header=T,na.strings=c("NA",""))
chimp_lengths<- fread("~/Desktop/Hi-C/gene_expression/chimp_lengths.txt", sep="\t",stringsAsFactors = FALSE,header=T,na.strings=c("NA",""))
#The function for RPKM conversion.
vRPKM <- function(expr.obj,chimp_lengths,human_lengths,meta4) {
if (is.null(expr.obj$E)) {
meta4%>%filter(SP=="C" & fullID %in% colnames(counts))->chimp_meta
meta4%>%filter(SP=="H" & fullID %in% colnames(counts))->human_meta
#using RPKM function:
#Put genes in correct order:
expr.obj$genes %>%select(Geneid=genes)%>%
left_join(.,chimp_lengths,by="Geneid")%>%select(Geneid,ch.length)->chlength
expr.obj$genes %>%select(Geneid=genes)%>%
left_join(.,human_lengths,by="Geneid")%>%select(Geneid,hu.length)->hulength
#Chimp RPKM
expr.obj$genes$Length<-(chlength$ch.length)
RPKMc=rpkm(expr.obj,normalized.lib.sizes=TRUE, log=TRUE)
RPKMc[,colnames(RPKMc) %in% chimp_meta$fullID]->rpkm_chimp
#Human RPKM
expr.obj$genes$Length<-hulength$hu.length
RPKMh=rpkm(expr.obj,normalized.lib.sizes=TRUE, log=TRUE)
RPKMh[,colnames(RPKMh) %in% human_meta$fullID]->rpkm_human
cbind(rpkm_chimp,rpkm_human)->allrpkm
expr.obj$E <- allrpkm
return(expr.obj)
}
else {
#Pull out gene order from voom object and add in gene lengths from feature counts file
#Put genes in correct order:
expr.obj$genes %>%select(Geneid=genes)%>%
left_join(.,chimp_lengths,by="Geneid")%>%select(Geneid,ch.length)->chlength
expr.obj$genes %>%select(Geneid=genes)%>%
left_join(.,human_lengths,by="Geneid")%>%select(Geneid,hu.length)->hulength
#Filter meta data to be able to separate human and chimp
meta4%>%filter(SP=="C")->chimp_meta
meta4%>%filter(SP=="H")->human_meta
#Pull out the expression data in cpm to convert to RPKM
expr.obj$E->forRPKM
forRPKM[,colnames(forRPKM) %in% chimp_meta$fullID]->rpkm_chimp
forRPKM[,colnames(forRPKM) %in% human_meta$fullID]->rpkm_human
#Make log2 in KB:
row.names(chlength)=chlength$Geneid
chlength %>% select(-Geneid)->chlength
as.matrix(chlength)->chlength
row.names(hulength)=hulength$Geneid
hulength %>% select(-Geneid)->hulength
as.matrix(hulength)->hulength
log2(hulength/1000)->l2hulength
log2(chlength/1000)->l2chlength
#Subtract out log2 kb:
sweep(rpkm_chimp, 1,l2chlength,"-")->chimp_rpkm
sweep(rpkm_human, 1,l2hulength,"-")->human_rpkm
colnames(forRPKM)->column_order
cbind(chimp_rpkm,human_rpkm)->vRPKMS
#Put RPKMS back into the VOOM object:
expr.obj$E <- (vRPKMS[,colnames(vRPKMS) %in% column_order])
return(expr.obj)
}
}
dge <- vRPKM(dge, chimp_lengths, human_lengths, group_ref) #Normalize via log2 RPKM.
#A typical low-expression filtering step: use default prior count adding (0.25), and filtering out anything that has fewer than half the individuals within each species having logCPM less than 1.5 (so want 2 humans AND 2 chimps with log2CPM >= 1.5)
lcpms <- cpm(dge$counts, log=TRUE) #Obtain log2CPM!
good.chimps <- which(rowSums(lcpms[,1:4]>=1.5)>=2) #Obtain good chimp indices
good.humans <- which(rowSums(lcpms[,5:8]>=1.5)>=2) #Obtain good human indices
filt <- good.humans[which(good.humans %in% good.chimps)] #Subsets us down to a solid 11,292 genes--will go for a similar percentage with RPKM cutoff vals! (25.6% of total)
#Repeat filtering step, this time on RPKMs. 0.4 was chosen as a cutoff as it obtains close to the same results as 1.5 lcpm (in terms of percentage of genes retained)
good.chimps <- which(rowSums(dge$E[,1:4]>=0.4)>=2) #Obtain good chimp indices.
good.humans <- which(rowSums(dge$E[,5:8]>=0.4)>=2) #Obtain good human indices.
RPKM_filt <- good.humans[which(good.humans %in% good.chimps)] #Still leaves us with 11,946 genes (27.1% of total)
#Do the actual filtering.
dge_filt <- dge[RPKM_filt,]
dge_filt$E <- dge$E[RPKM_filt,]
dge_filt$counts <- dge$counts[RPKM_filt,]
dge_final <- calcNormFactors(dge_filt, method="TMM") #Calculate normalization factors with trimmed mean of M-values (TMM).
dge_norm <- calcNormFactors(dge, method="TMM") #Calculate normalization factors with TMM on dge before filtering out lowly expressed genes, for normalization visualization.
#Quick visualization of the filtering I've just performed:
col <- brewer.pal(8, "Paired")
par(mfrow=c(1,2))
plot(density(dge$E[,1]), col=col[1], lwd=2, ylim=c(0,0.35), las=2,
main="", xlab="")
title(main="A. Raw data", xlab="Log-RPKM")
abline(v=1, lty=3)
for (i in 2:8){
den <- density(dge$E[,i])
lines(den$x, den$y, col=col[i], lwd=2)
}
legend("topright", colnames(dge$E[,1:8]), text.col=col, bty="n")
plot(density(dge_final$E[,1]), col=col[1], lwd=2, ylim=c(0,0.35), las=2,
main="", xlab="")
title(main="B. Filtered data", xlab="Log-RPKM")
abline(v=1, lty=3)
for (i in 2:8){
den <- density(dge_final$E[,i])
lines(den$x, den$y, col=col[i], lwd=2)
}
legend("topright", colnames(dge_final[,1:8]), text.col=col, bty="n")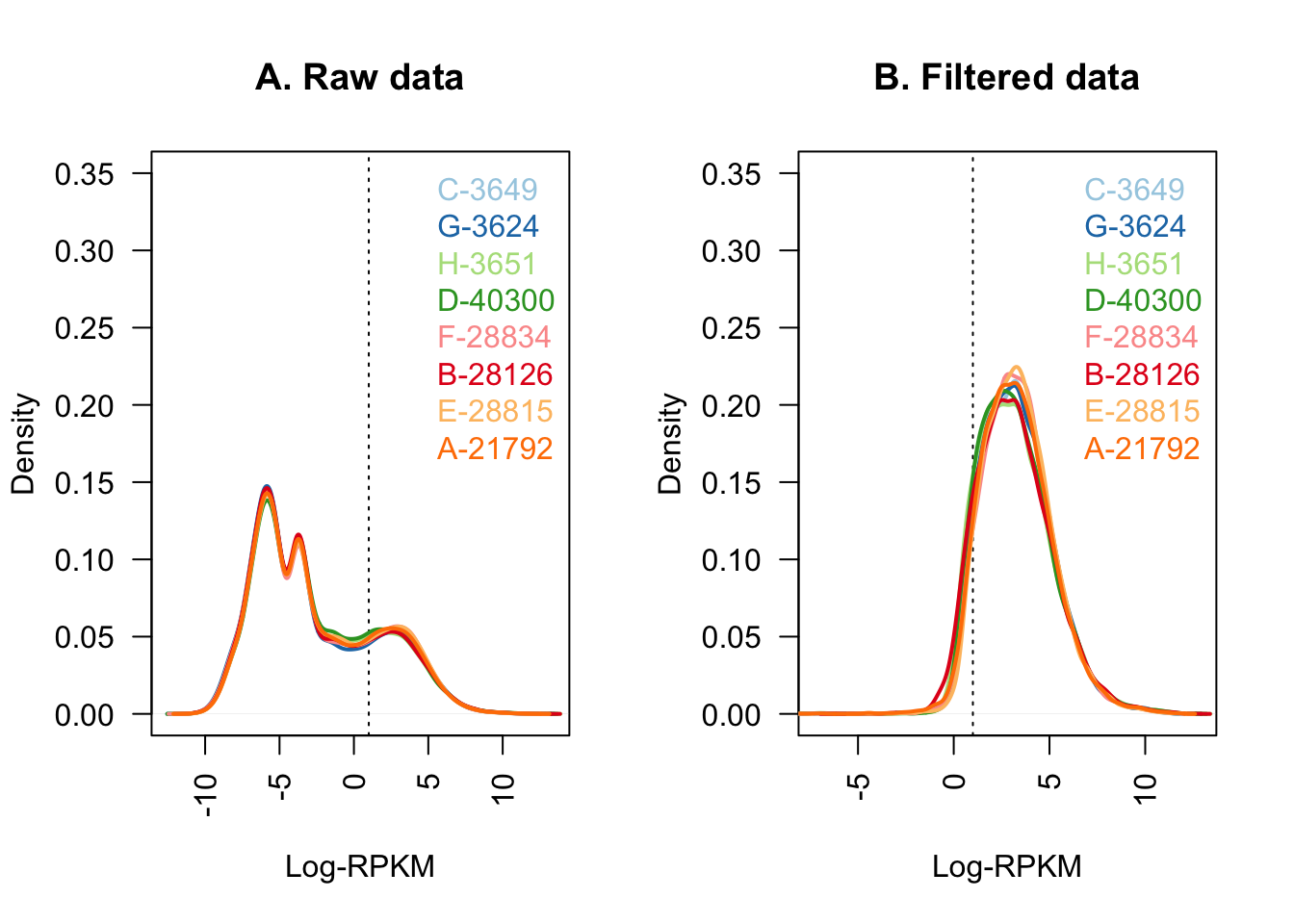
#Quick visualization of the normalization on the whole set of genes.
col <- brewer.pal(8, "Paired")
raw <- as.data.frame(dge$E[,1:8])
normed <- as.data.frame(dge_norm$E[,1:8])
par(mfrow=c(1,2))
boxplot(raw, las=2, col=col, main="")
title(main="Unnormalized data",ylab="Log-RPKM")
boxplot(normed, las=2, col=col, main="")
title(main="Normalized data",ylab="Log-RPKM")
#Now, observe normalization on the filtered set of genes.
col <- brewer.pal(8, "Paired")
raw <- as.data.frame(dge_filt$E[,1:8])
normed <- as.data.frame(dge_final$E[,1:8])
par(mfrow=c(1,2))
boxplot(raw, las=2, col=col, main="")
title(main="Unnormalized data",ylab="Log-RPKM")
boxplot(normed, las=2, col=col, main="")
title(main="Normalized data",ylab="Log-RPKM")
#Now, do some quick MDS plotting to make sure this expression data separates out species properly.
species <- c("C", "C", "C", "C", "H", "H", "H", "H")
color <- c(rep("red", 4), rep("blue", 4))
par(mfrow=c(1,1))
plotMDS(dge_final$E[,1:8], labels=species, col=color, main="MDS Plot") #Shows separation of the species along the logFC dimension representing the majority of the variance--orthogonal check to PCA, and looks great!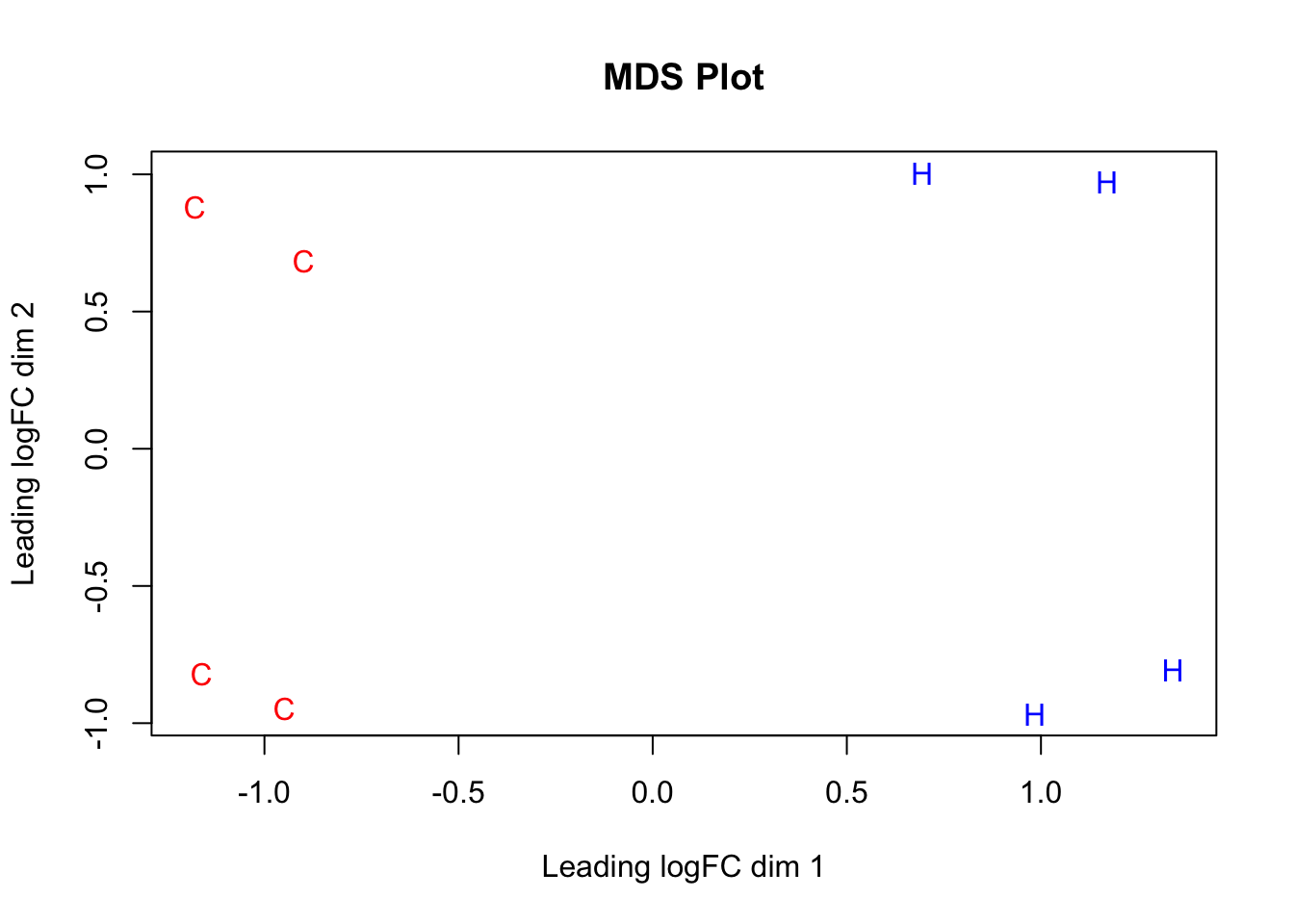
###Now, apply voom to get quality weights.
meta.exp.data <- data.frame("SP"=c("C", "C", "C", "C", "H", "H", "H", "H"), "SX"=c("M","M" ,"F","F","F", "M","M","F"))
SP <- factor(meta.exp.data$SP,levels = c("H","C"))
exp.design <- model.matrix(~0+SP)#(~1+meta.exp.data$SP+meta.exp.data$SX)
colnames(exp.design) <- c("Human", "Chimp")
weighted.data <- voom(dge_final, exp.design, plot=TRUE, normalize.method = "cyclicloess")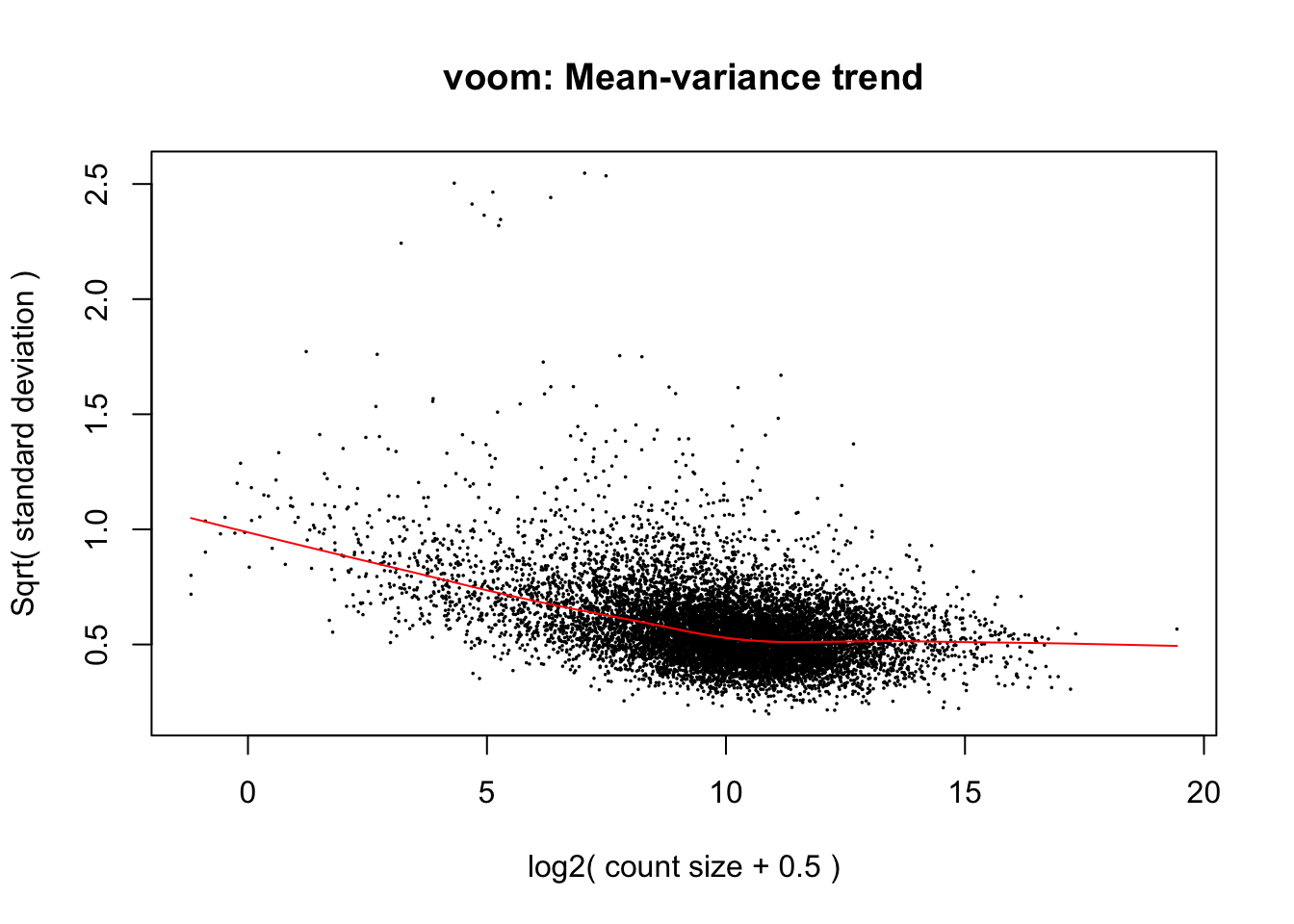
##Obtain rest of LM results, with particular eye to DE table!
vfit <- lmFit(weighted.data, exp.design)
efit <- eBayes(vfit)
mycon <- makeContrasts(HvC = Human-Chimp, levels = exp.design)
diff_species <- contrasts.fit(efit, mycon)
finalfit <- eBayes(diff_species)
detable <- topTable(finalfit, coef = 1, adjust.method = "BH", number = Inf, sort.by="none")
plotSA(efit, main="Final model: Mean−variance trend")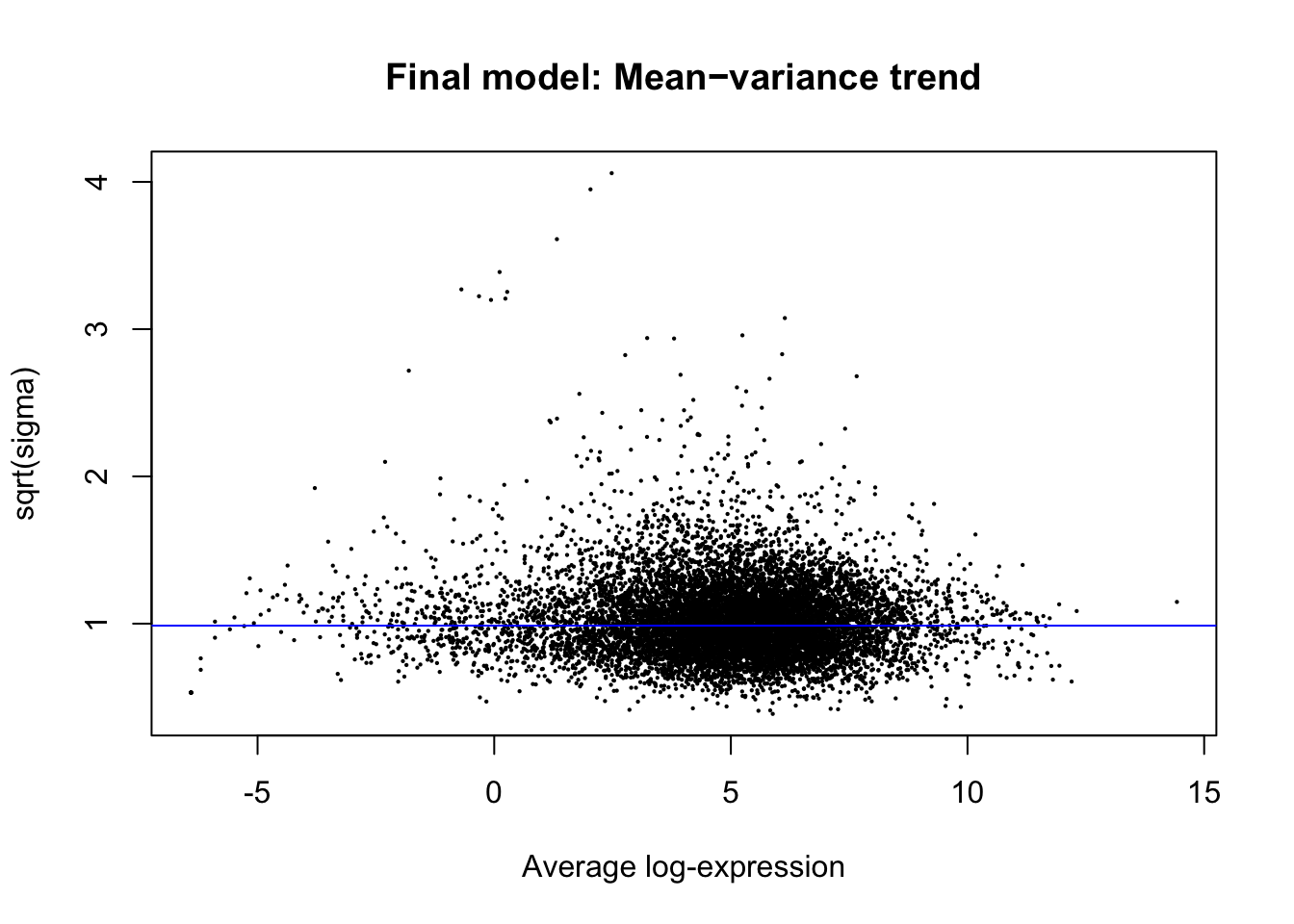
#Get lists of the DE and non-DE genes so I can run separate analyses on them at any point.
DEgenes <- detable$genes[which(detable$adj.P.Val<=0.05)]
nonDEgenes <- detable$genes[-which(detable$adj.P.Val<=0.05)]
#Rearrange RPKM and weight columns in voom object to be similar to the rest of my setup throughout in other dataframes.
weighted.data$E <- weighted.data$E[,c(8, 6, 1, 4, 7, 5, 2, 3)]
weighted.data$weights <- weighted.data$weights[,c(8, 6, 1, 4, 7, 5, 2, 3)]
RPKM <- weighted.data$E
rownames(RPKM) <- NULL #Just to match what I had before on midway2, about to write this out.
saveRDS(RPKM, file="~/Desktop/Hi-C/HiC_covs/IEE.RPKM.RDS")
saveRDS(weighted.data, file="~/Desktop/Hi-C/HiC_covs/IEE_voom_object.RDS") #write this object out, can then be read in with readRDS.Overlap Between Juicer Data and Orthogonal Gene Expression Data
In this section I find the overlap between the 2 final filtered set of Hi-C Juicer significant hits and genes picked up on by an orthogonal RNA-seq experiment in the same set of cell lines. I utilize an in-house curated set of orthologous genes between humans and chimpanzees. Given that the resolution of the data is 10kb, I choose a simple and conservative approach and use a 1-nucleotide interval at the start of each gene as a proxy for the promoter. I then take a conservative pass and only use genes that had direct overlap with a bin from the Hi-C significant hits data, with more motivation explained below.
#Now, read in filtered data from linear_modeling_QC.Rmd.
data.KR <- fread("~/Desktop/Hi-C/juicer.filt.KR.final", header=TRUE, data.table=FALSE, stringsAsFactors = FALSE, showProgress=FALSE)
data.VC <- fread("~/Desktop/Hi-C/juicer.filt.VC.final", header=TRUE, data.table=FALSE, stringsAsFactors = FALSE, showProgress = FALSE)
meta.data <- data.frame("SP"=c("H", "H", "C", "C", "H", "H", "C", "C"), "SX"=c("F", "M", "M", "F", "M", "F", "M", "F"), "Batch"=c(1, 1, 1, 1, 2, 2, 2, 2))
#####GENE Hi-C Hit overlap: Already have hgenes and cgenes properly formatted and in the necessary folder from the HOMER version of this analysis. See HOMER version of gene_expression analysis for more details on how I obtained orthologous genes.
#Now, what I will need to do is prep bed files from the 2 sets of filtered data for each bin, in order to run bedtools-closest on them with the human and chimp gene data. This is for getting each bin's proximity to TSS by overlapping with the dfs I was just referring to (humgenes and chimpgenes). In the end this set of bedfiles is fairly useless, because really it would be preferable to get rid of duplicates so that I can merely group_by on a given bin afterwards and left_join as necessary. So somewhat deprecated, but I keep it here still:
hbin1KR <- data.frame(chr=data.KR$Hchr, start=as.numeric(gsub("chr.*-", "", data.KR$H1)), end=as.numeric(gsub("chr.*-", "", data.KR$H1))+10000)
hbin2KR <- data.frame(chr=data.KR$Hchr, start=as.numeric(gsub("chr.*-", "", data.KR$H2)), end=as.numeric(gsub("chr.*-", "", data.KR$H2))+10000)
cbin1KR <- data.frame(chr=data.KR$Cchr, start=as.numeric(gsub("chr.*-", "", data.KR$C1)), end=as.numeric(gsub("chr.*-", "", data.KR$C1))+10000)
cbin2KR <- data.frame(chr=data.KR$Cchr, start=as.numeric(gsub("chr.*-", "", data.KR$C2)), end=as.numeric(gsub("chr.*-", "", data.KR$C2))+10000)
hbin1VC <- data.frame(chr=data.VC$Hchr, start=as.numeric(gsub("chr.*-", "", data.VC$H1)), end=as.numeric(gsub("chr.*-", "", data.VC$H1))+10000)
hbin2VC <- data.frame(chr=data.VC$Hchr, start=as.numeric(gsub("chr.*-", "", data.VC$H2)), end=as.numeric(gsub("chr.*-", "", data.VC$H2))+10000)
cbin1VC <- data.frame(chr=data.VC$Cchr, start=as.numeric(gsub("chr.*-", "", data.VC$C1)), end=as.numeric(gsub("chr.*-", "", data.VC$C1))+10000)
cbin2VC <- data.frame(chr=data.VC$Cchr, start=as.numeric(gsub("chr.*-", "", data.VC$C2)), end=as.numeric(gsub("chr.*-", "", data.VC$C2))+10000)
#In most analyses, it will make more sense to have a single bed file for both sets of bins, and remove all duplicates. I create that here:
hbinsKR <- rbind(hbin1KR[!duplicated(hbin1KR),], hbin2KR[!duplicated(hbin2KR),])
hbinsKR <- hbinsKR[!duplicated(hbinsKR),]
cbinsKR <- rbind(cbin1KR[!duplicated(cbin1KR),], cbin2KR[!duplicated(cbin2KR),])
cbinsKR <- cbinsKR[!duplicated(cbinsKR),]
hbinsVC <- rbind(hbin1VC[!duplicated(hbin1VC),], hbin2VC[!duplicated(hbin2VC),])
hbinsVC <- hbinsVC[!duplicated(hbinsVC),]
cbinsVC <- rbind(cbin1VC[!duplicated(cbin1VC),], cbin2VC[!duplicated(cbin2VC),])
cbinsVC <- cbinsVC[!duplicated(cbinsVC),]
#Now, write all of these files out for analysis with bedtools.
options(scipen=999) #Don't want any scientific notation in these BED files, will mess up some bedtools analyses at times
write.table(hbin1KR, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/hbin1KR.bed", quote = FALSE, sep="\t", row.names = FALSE, col.names=FALSE)
write.table(hbin2KR, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/hbin2KR.bed", quote = FALSE, sep="\t", row.names = FALSE, col.names=FALSE)
write.table(cbin1KR, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/cbin1KR.bed", quote = FALSE, sep="\t", row.names = FALSE, col.names=FALSE)
write.table(cbin2KR, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/cbin2KR.bed", quote = FALSE, sep="\t", row.names = FALSE, col.names=FALSE)
write.table(hbinsKR, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/hbinsKR.bed", quote=FALSE, sep="\t", row.names=FALSE, col.names=FALSE)
write.table(cbinsKR, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/cbinsKR.bed", quote=FALSE, sep="\t", row.names=FALSE, col.names=FALSE)
write.table(hbin1VC, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/hbin1VC.bed", quote = FALSE, sep="\t", row.names = FALSE, col.names=FALSE)
write.table(hbin2VC, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/hbin2VC.bed", quote = FALSE, sep="\t", row.names = FALSE, col.names=FALSE)
write.table(cbin1VC, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/cbin1VC.bed", quote = FALSE, sep="\t", row.names = FALSE, col.names=FALSE)
write.table(cbin2VC, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/cbin2VC.bed", quote = FALSE, sep="\t", row.names = FALSE, col.names=FALSE)
write.table(hbinsVC, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/hbinsVC.bed", quote=FALSE, sep="\t", row.names=FALSE, col.names=FALSE)
write.table(cbinsVC, "~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/unsorted/cbinsVC.bed", quote=FALSE, sep="\t", row.names=FALSE, col.names=FALSE)
options(scipen=0)
#Read in new, simpler bedtools closest files for genes. This is after running two commands, after sorting the files w/ sort -k1,1 -k2,2n in.bed > out.bed:
#bedtools closest -D a -a cgenes.sorted.bed -b cbins.sorted.bed > cgene.hic.overlap
#bedtools closest -D a -a hgenes.sorted.bed -b hbins.sorted.bed > hgene.hic.overlap
hgene.hic.KR <- fread("~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/hgene.hic.KR.overlap", header=FALSE, stringsAsFactors = FALSE, data.table=FALSE)
cgene.hic.KR <- fread("~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/cgene.hic.KR.overlap", header=FALSE, stringsAsFactors = FALSE, data.table=FALSE)
hgene.hic.VC <- fread("~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/hgene.hic.VC.overlap", header=FALSE, stringsAsFactors = FALSE, data.table=FALSE)
cgene.hic.VC <- fread("~/Desktop/Hi-C/gene_expression/juicer_10kb_filt_overlaps/cgene.hic.VC.overlap", header=FALSE, stringsAsFactors = FALSE, data.table=FALSE)
#Visualize the overlap of genes with bins and see how many genes we get back! Left out here b/c not super necessary, but code is collapsed here:
# hum.genelap <- data.frame(overlap=seq(0, 100000, 1000), perc.genes = NA, tot.genes=NA)
# for(row in 1:nrow(hum.genelap)){
# hum.genelap$perc.genes[row] <- sum(abs(hgene.hic$V10)<=hum.genelap$overlap[row])/length(hgene.hic$V10)
# hum.genelap$tot.genes[row] <- sum(abs(hgene.hic$V10)<=hum.genelap$overlap[row])
# }
#
# c.genelap <- data.frame(overlap=seq(0, 100000, 1000), perc.genes=NA, tot.genes=NA)
# for(row in 1:nrow(c.genelap)){
# c.genelap$perc.genes[row] <- sum(abs(cgene.hic$V10)<=c.genelap$overlap[row])/length(cgene.hic$V10)
# c.genelap$tot.genes[row] <- sum(abs(cgene.hic$V10)<=c.genelap$overlap[row])
# }
# c.genelap$type <- "chimp"
# hum.genelap$type <- "human"
#Examine what the potential gains are here if we are more lenient about the overlap/closeness to a TSS...
#ggoverlap <- rbind(hum.genelap, c.genelap)
#ggplot(data=ggoverlap) + geom_line(aes(x=overlap, y=perc.genes*100, color=type)) + ggtitle("Percent of Total Genes Picked Up | Min. Distance from TSS") + xlab("Distance from TSS") + ylab("Percentage of genes in ortho exon trios file (~44k)") + scale_color_discrete(guide=guide_legend(title="Species")) + coord_cartesian(xlim=c(0, 30000)) + scale_x_continuous(breaks=seq(0, 30000, 5000))
#ggplot(data=ggoverlap) + geom_line(aes(x=overlap, y=tot.genes, color=type)) + ggtitle("Total # Genes Picked Up | Min. Distance from TSS") + xlab("Distance from TSS") + ylab("Total # of Genes Picked up On (of ~44k)") + scale_color_discrete(guide=guide_legend(title="Species")) + coord_cartesian(xlim=c(0, 30000)) + scale_x_continuous(breaks=seq(0, 30000, 5000))
#Start with a conservative pass--only take those genes that had an actual overlap with a bin, not ones that were merely close to one. Allowing some leeway to include genes that are within 1kb, 2kb, 3kb etc. of a Hi-C bin adds an average of ~800 genes per 1kb. We can also examine the distribution manually to motivate this decision:
quantile(abs(hgene.hic.KR$V10), probs=seq(0, 1, 0.025)) 0% 2.5% 5% 7.5% 10% 12.5% 15%
0.0 0.0 0.0 0.0 1.0 1.0 111.2
17.5% 20% 22.5% 25% 27.5% 30% 32.5%
2801.8 5837.6 9170.0 12712.0 16555.3 20657.4 25122.3
35% 37.5% 40% 42.5% 45% 47.5% 50%
30415.8 35854.0 41976.0 48686.7 56315.6 64775.6 74086.0
52.5% 55% 57.5% 60% 62.5% 65% 67.5%
84493.0 96003.4 109333.4 124395.4 141705.5 161762.6 185550.4
70% 72.5% 75% 77.5% 80% 82.5% 85%
213137.8 245623.6 284011.0 326249.0 378968.0 446350.3 531628.2
87.5% 90% 92.5% 95% 97.5% 100%
644495.0 813443.4 1066258.7 1411418.6 2117706.0 8009993.0 quantile(abs(cgene.hic.KR$V10), probs=seq(0, 1, 0.025)) 0% 2.5% 5% 7.5% 10% 12.5% 15%
0.0 0.0 0.0 0.0 1.0 1.0 35.8
17.5% 20% 22.5% 25% 27.5% 30% 32.5%
2576.0 5543.2 9001.0 12613.0 16346.1 20659.2 25304.9
35% 37.5% 40% 42.5% 45% 47.5% 50%
30415.0 36090.5 42336.4 49350.3 57203.8 65644.9 74758.0
52.5% 55% 57.5% 60% 62.5% 65% 67.5%
85634.5 97537.4 110882.8 125779.8 143152.5 163577.6 187829.8
70% 72.5% 75% 77.5% 80% 82.5% 85%
216384.2 248610.8 288103.0 330921.3 384602.4 448229.0 536251.6
87.5% 90% 92.5% 95% 97.5% 100%
645908.0 806354.8 1046000.8 1398275.0 2071010.3 7529283.0 quantile(abs(hgene.hic.VC$V10), probs=seq(0, 1, 0.025)) 0% 2.5% 5% 7.5% 10% 12.5% 15%
0.0 0.0 0.0 0.0 0.0 748.5 2939.6
17.5% 20% 22.5% 25% 27.5% 30% 32.5%
5319.8 8011.8 10835.5 13955.0 16964.2 20339.0 24054.6
35% 37.5% 40% 42.5% 45% 47.5% 50%
28368.6 33051.5 38083.0 43953.0 50389.2 57444.3 64971.0
52.5% 55% 57.5% 60% 62.5% 65% 67.5%
73526.5 83118.4 94055.9 106232.2 119220.5 135994.6 153922.6
70% 72.5% 75% 77.5% 80% 82.5% 85%
175048.2 199192.6 228645.0 261707.1 302957.2 351916.0 413198.4
87.5% 90% 92.5% 95% 97.5% 100%
501881.5 641720.2 865347.1 1205121.8 1836870.5 6456300.0 quantile(abs(cgene.hic.VC$V10), probs=seq(0, 1, 0.025)) 0% 2.5% 5% 7.5% 10% 12.5% 15%
0.0 0.0 0.0 0.0 0.0 512.5 2649.0
17.5% 20% 22.5% 25% 27.5% 30% 32.5%
5010.2 7760.8 10585.5 13538.0 16670.4 20081.6 24187.6
35% 37.5% 40% 42.5% 45% 47.5% 50%
28398.8 33210.5 38408.6 44155.4 50593.8 57687.0 65471.0
52.5% 55% 57.5% 60% 62.5% 65% 67.5%
73931.1 83568.4 94835.9 107072.8 120344.5 136233.2 154759.7
70% 72.5% 75% 77.5% 80% 82.5% 85%
176847.0 202064.3 230537.0 264555.9 303717.0 353338.0 415156.6
87.5% 90% 92.5% 95% 97.5% 100%
501151.5 629124.0 840981.2 1183185.6 1767476.4 6500471.0 #So it looks as though we pick up on approximately 10% of 44k genes (~4.4k)
#Note I looked at proportion of overlap with DE and with non-DE genes just for curiosity, and roughly 66% of the DE genes have overlap with a Hi-C bin while roughly 70% of the non-DE genes do. Since this result isn't particularly interesting I have collapsed that analysis here.
#Also are interested in seeing how this differs for DE and non-DE genes.
sum(hgene.hic.KR$V10==0&(hgene.hic.KR$V4 %in% DEgenes)) #263 genes[1] 263sum(hgene.hic.KR$V10==0&(hgene.hic.KR$V4 %in% nonDEgenes)) #1011 genes[1] 1011sum(hgene.hic.KR$V10==0&(!hgene.hic.KR$V4 %in% DEgenes)&(!hgene.hic.KR$V4 %in% nonDEgenes)) #Checking which genes have direct overlap with bins, but were not picked up on in our RNAseq data. 2717[1] 2717sum(hgene.hic.VC$V10==0&(hgene.hic.VC$V4 %in% DEgenes)) #334 genes[1] 334sum(hgene.hic.VC$V10==0&(hgene.hic.VC$V4 %in% nonDEgenes)) #1296 genes[1] 1296sum(hgene.hic.VC$V10==0&(!hgene.hic.VC$V4 %in% DEgenes)&(!hgene.hic.VC$V4 %in% nonDEgenes)) #Checking which genes have direct overlap with bins, but were not picked up on in our RNAseq data. 3441[1] 3441Linear Modeling Annotation
In this next section I simply add information obtained from linear modeling on the Hi-C interaction frequencies to the appropriate genes having overlap with Hi-C bins. Because one Hi-C bin frequently shows up many times in the data, this means I must choose some kind of summary for Hi-C contact frequencies and linear modeling annotations for each gene. I toy with a variety of these summaries here, including choosing the minimum FDR contact, the maximum beta contact, the upstream contact, or summarizing all a bin’s contacts with the weighted Z-combine method or median FDR values.
hgene.hic.overlap.KR <- filter(hgene.hic.KR, V10==0) #Still leaves a solid ~4k genes.
cgene.hic.overlap.KR <- filter(cgene.hic.KR, V10==0) #Still leaves a solid ~4k genes.
hgene.hic.overlap.VC <- filter(hgene.hic.VC, V10==0) #Still leaves a solid ~5k genes.
cgene.hic.overlap.VC <- filter(cgene.hic.VC, V10==0) #Still leaves a solid ~5k genes.
#Add a column to both dfs indicating where along a bin the gene in question is found (from 0-10k):
hgene.hic.overlap.KR$bin_pos <- abs(hgene.hic.overlap.KR$V8-hgene.hic.overlap.KR$V2)
cgene.hic.overlap.KR$bin_pos <- abs(cgene.hic.overlap.KR$V8-cgene.hic.overlap.KR$V2)
hgene.hic.overlap.VC$bin_pos <- abs(hgene.hic.overlap.VC$V8-hgene.hic.overlap.VC$V2)
cgene.hic.overlap.VC$bin_pos <- abs(cgene.hic.overlap.VC$V8-cgene.hic.overlap.VC$V2)
#Rearrange columns and create another column of the bin ID.
hgene.hic.overlap.KR <- hgene.hic.overlap.KR[,c(4, 7:9, 6, 11, 1:2)]
hgene.hic.overlap.KR$HID <- paste(hgene.hic.overlap.KR$V7, hgene.hic.overlap.KR$V8, sep="-")
cgene.hic.overlap.KR <- cgene.hic.overlap.KR[,c(4, 7:9, 6, 11, 1:2)]
cgene.hic.overlap.KR$CID <- paste(cgene.hic.overlap.KR$V7, cgene.hic.overlap.KR$V8, sep="-")
colnames(hgene.hic.overlap.KR) <- c("genes", "HiC_chr", "H1start", "H1end", "Hstrand", "bin_pos", "genechr", "genepos", "HID")
colnames(cgene.hic.overlap.KR) <- c("genes", "HiC_chr", "C1start", "C1end", "Cstrand", "bin_pos", "genechr", "genepos", "CID")
hgene.hic.overlap.VC <- hgene.hic.overlap.VC[,c(4, 7:9, 6, 11, 1:2)]
hgene.hic.overlap.VC$HID <- paste(hgene.hic.overlap.VC$V7, hgene.hic.overlap.VC$V8, sep="-")
cgene.hic.overlap.VC <- cgene.hic.overlap.VC[,c(4, 7:9, 6, 11, 1:2)]
cgene.hic.overlap.VC$CID <- paste(cgene.hic.overlap.VC$V7, cgene.hic.overlap.VC$V8, sep="-")
colnames(hgene.hic.overlap.VC) <- c("genes", "HiC_chr", "H1start", "H1end", "Hstrand", "bin_pos", "genechr", "genepos", "HID")
colnames(cgene.hic.overlap.VC) <- c("genes", "HiC_chr", "C1start", "C1end", "Cstrand", "bin_pos", "genechr", "genepos", "CID")
#Before extracting some data from the data.KR and data.VC dfs, need to reformat their H1, H2, C1, and C2 columns to include chromosome (this was how homer data already was formatted so didn't have to think about it there)
data.KR$H1 <- paste(data.KR$Hchr, data.KR$H1, sep="-")
data.KR$H2 <- paste(data.KR$Hchr, data.KR$H2, sep="-")
data.VC$H1 <- paste(data.VC$Hchr, data.VC$H1, sep="-")
data.VC$H2 <- paste(data.VC$Hchr, data.VC$H2, sep="-")
data.KR$C1 <- paste(data.KR$Cchr, data.KR$C1, sep="-")
data.KR$C2 <- paste(data.KR$Cchr, data.KR$C2, sep="-")
data.VC$C1 <- paste(data.VC$Cchr, data.VC$C1, sep="-")
data.VC$C2 <- paste(data.VC$Cchr, data.VC$C2, sep="-")
hbindf.KR <- select(data.KR, "H1", "H2", "ALLvar", "SE", "sp_beta", "sp_pval", "sp_BH_pval", "Hdist")
names(hbindf.KR) <- c("HID", "HID2", "ALLvar", "SE", "sp_beta", "sp_pval", "sp_BH_pval", "distance") #I have confirmed that all the HID2s are higher numbered coordinates than the HID1s, the only instance in which this isn't the case is when the two bins are identical (this should have been filtered out long before now).
hbindf.VC <- select(data.VC, "H1", "H2", "ALLvar", "SE", "sp_beta", "sp_pval", "sp_BH_pval", "Hdist")
names(hbindf.VC) <- c("HID", "HID2", "ALLvar", "SE", "sp_beta", "sp_pval", "sp_BH_pval", "distance")
#This step is unnecessary in this paradigm as there are no hits like this, but keep it in here for the premise's sake:
hbindf.KR <- hbindf.KR[(which(hbindf.KR$distance!=0)),] #Removes pairs where the same bin represents both mates. These instances occur exclusively when liftOver of the genomic coordinates from one species to another, and the subsequent rounding to the nearest 10kb, results in a contact between adjacent bins in one species being mapped as a contact between the same bin in the other species. Because there are less than 50 instances of this total in the dataset I simply remove it here without further worry.
hbindf.VC <- hbindf.VC[(which(hbindf.VC$distance!=0)),]
#For explanations about the motivations and intents behind code in the rest of this chunk, please see the same file for the HOMER analysis.
group_by(hbindf.KR, HID) %>% summarise(DS_bin=HID2[which.min(distance)], DS_FDR=sp_BH_pval[which.min(distance)], DS_dist=distance[which.min(distance)]) -> hbin1.downstream.KR
group_by(hbindf.KR, HID2) %>% summarise(US_bin=HID[which.min(distance)], US_FDR=sp_BH_pval[which.min(distance)], US_dist=distance[which.min(distance)]) -> hbin2.upstream.KR
colnames(hbin2.upstream.KR) <- c("HID", "US_bin", "US_FDR", "US_dist")
Hstreams.KR <- full_join(hbin1.downstream.KR, hbin2.upstream.KR, by="HID")
hbindf.KR.flip <- hbindf.KR[,c(2, 1, 3:7)]
colnames(hbindf.KR.flip)[1:2] <- c("HID", "HID2")
hbindf.KR_x2 <- rbind(hbindf.KR[,1:7], hbindf.KR.flip)
as.data.frame(group_by(hbindf.KR_x2, HID) %>% summarise(min_FDR_bin=HID2[which.min(sp_BH_pval)], min_FDR=min(sp_BH_pval), min_FDR_pval=sp_pval[which.min(sp_BH_pval)], min_FDR_B=sp_beta[which.min(sp_BH_pval)], median_FDR=median(sp_BH_pval), weighted_Z.ALLvar=pnorm((sum((1/ALLvar)*((qnorm(1-sp_pval))))/sqrt(sum((1/ALLvar)^2))), lower.tail=FALSE), weighted_Z.s2post=pnorm(sum((1/(SE^2))*qnorm(1-sp_pval))/sqrt(sum(1/SE^2)), lower.tail=FALSE), fisher=-2*sum(log(sp_pval)), numcontacts=n(), max_B_bin=HID2[which.max(abs(sp_beta))], max_B_FDR=sp_BH_pval[which.max(abs(sp_beta))], max_B=sp_beta[which.max(abs(sp_beta))])) -> hbin.KR.info
full_join(hbin.KR.info, Hstreams.KR, by="HID") -> hbin.KR.full.info
left_join(hgene.hic.overlap.KR, hbin.KR.full.info, by="HID") -> humgenes.KR.full
colnames(humgenes.KR.full)[1:5] <- c("genes", "Hchr", "Hstart", "Hend", "Hstrand") #Fix column names for what was just created
group_by(hbindf.VC, HID) %>% summarise(DS_bin=HID2[which.min(distance)], DS_FDR=sp_BH_pval[which.min(distance)], DS_dist=distance[which.min(distance)]) -> hbin1.downstream.VC
group_by(hbindf.VC, HID2) %>% summarise(US_bin=HID[which.min(distance)], US_FDR=sp_BH_pval[which.min(distance)], US_dist=distance[which.min(distance)]) -> hbin2.upstream.VC
colnames(hbin2.upstream.VC) <- c("HID", "US_bin", "US_FDR", "US_dist")
Hstreams.VC <- full_join(hbin1.downstream.VC, hbin2.upstream.VC, by="HID")
hbindf.VC.flip <- hbindf.VC[,c(2, 1, 3:7)]
colnames(hbindf.VC.flip)[1:2] <- c("HID", "HID2")
hbindf.VC_x2 <- rbind(hbindf.VC[,1:7], hbindf.VC.flip)
as.data.frame(group_by(hbindf.VC_x2, HID) %>% summarise(min_FDR_bin=HID2[which.min(sp_BH_pval)], min_FDR=min(sp_BH_pval), min_FDR_pval=sp_pval[which.min(sp_BH_pval)], min_FDR_B=sp_beta[which.min(sp_BH_pval)], median_FDR=median(sp_BH_pval), weighted_Z.ALLvar=pnorm((sum((1/ALLvar)*((qnorm(1-sp_pval))))/sqrt(sum((1/ALLvar)^2))), lower.tail=FALSE), weighted_Z.s2post=pnorm(sum((1/(SE^2))*qnorm(1-sp_pval))/sqrt(sum(1/SE^2)), lower.tail=FALSE), fisher=-2*sum(log(sp_pval)), numcontacts=n(), max_B_bin=HID2[which.max(abs(sp_beta))], max_B_FDR=sp_BH_pval[which.max(abs(sp_beta))], max_B=sp_beta[which.max(abs(sp_beta))])) -> hbin.VC.info
full_join(hbin.VC.info, Hstreams.VC, by="HID") -> hbin.VC.full.info
left_join(hgene.hic.overlap.VC, hbin.VC.full.info, by="HID") -> humgenes.VC.full
colnames(humgenes.VC.full)[1:5] <- c("genes", "Hchr", "Hstart", "Hend", "Hstrand") #Fix column names for what was just created
###Do chimps in both as well, to maximize overlaps:
###KR
cbin.KR <- select(data.KR, "C1", "C2", "ALLvar", "SE", "sp_beta", "sp_pval", "sp_BH_pval", "Cdist")
names(cbin.KR) <- c("CID", "CID2", "ALLvar", "SE", "sp_beta", "sp_pval", "sp_BH_pval", "distance")
cbin.KR <- cbin.KR[(which(cbin.KR$dist!=0)),]
group_by(cbin.KR, CID) %>% summarise(DS_bin=CID2[which.min(distance)], DS_FDR=sp_BH_pval[which.min(distance)], DS_dist=distance[which.min(distance)]) -> cbin1.downstream.KR
group_by(cbin.KR, CID2) %>% summarise(US_bin=CID[which.min(distance)], US_FDR=sp_BH_pval[which.min(distance)], US_dist=distance[which.min(distance)]) -> cbin2.upstream.KR
colnames(cbin2.upstream.KR) <- c("CID", "US_bin", "US_FDR", "US_dist")
Cstreams.KR <- full_join(cbin1.downstream.KR, cbin2.upstream.KR, by="CID")
cbin.KR.flip <- cbin.KR[,c(2, 1, 3:7)]
colnames(cbin.KR.flip)[1:2] <- c("CID", "CID2")
cbin.KR_x2 <- rbind(cbin.KR[,1:7], cbin.KR.flip)
group_by(cbin.KR_x2, CID) %>% summarise(min_FDR_bin=CID2[which.min(sp_BH_pval)], min_FDR=min(sp_BH_pval), min_FDR_B=sp_beta[which.min(sp_BH_pval)], median_FDR=median(sp_BH_pval), weighted_Z.ALLvar=pnorm((sum((1/ALLvar)*((qnorm(1-sp_pval))))/sqrt(sum((1/ALLvar)^2))), lower.tail=FALSE), weighted_Z.s2post=pnorm(sum((1/(SE^2))*qnorm(1-sp_pval))/sqrt(sum(1/SE^2)), lower.tail=FALSE), fisher=-2*sum(log(sp_pval)), numcontacts=n(), max_B_bin=CID2[which.max(abs(sp_beta))], max_B_FDR=sp_BH_pval[which.max(abs(sp_beta))], max_B=sp_beta[which.max(abs(sp_beta))]) -> cbin.info.KR
full_join(cbin.info.KR, Cstreams.KR, by="CID") -> cbin.full.info.KR
left_join(cgene.hic.overlap.KR, cbin.full.info.KR, by="CID") -> chimpgenes.hic.KR
colnames(chimpgenes.hic.KR)[1:5] <- c("genes", "Hchr", "Hstart", "Hend", "Hstrand") #Fix column names for what was just created
###VC
cbin.VC <- select(data.VC, "C1", "C2", "ALLvar", "SE", "sp_beta", "sp_pval", "sp_BH_pval", "Cdist")
names(cbin.VC) <- c("CID", "CID2", "ALLvar", "SE", "sp_beta", "sp_pval", "sp_BH_pval", "distance")
cbin.VC <- cbin.VC[(which(cbin.VC$dist!=0)),]
group_by(cbin.VC, CID) %>% summarise(DS_bin=CID2[which.min(distance)], DS_FDR=sp_BH_pval[which.min(distance)], DS_dist=distance[which.min(distance)]) -> cbin1.downstream.VC
group_by(cbin.VC, CID2) %>% summarise(US_bin=CID[which.min(distance)], US_FDR=sp_BH_pval[which.min(distance)], US_dist=distance[which.min(distance)]) -> cbin2.upstream.VC
colnames(cbin2.upstream.VC) <- c("CID", "US_bin", "US_FDR", "US_dist")
Cstreams.VC <- full_join(cbin1.downstream.VC, cbin2.upstream.VC, by="CID")
cbin.VC.flip <- cbin.VC[,c(2, 1, 3:7)]
colnames(cbin.VC.flip)[1:2] <- c("CID", "CID2")
cbin.VC_x2 <- rbind(cbin.VC[,1:7], cbin.VC.flip)
group_by(cbin.VC_x2, CID) %>% summarise(min_FDR_bin=CID2[which.min(sp_BH_pval)], min_FDR=min(sp_BH_pval), min_FDR_B=sp_beta[which.min(sp_BH_pval)], median_FDR=median(sp_BH_pval), weighted_Z.ALLvar=pnorm((sum((1/ALLvar)*((qnorm(1-sp_pval))))/sqrt(sum((1/ALLvar)^2))), lower.tail=FALSE), weighted_Z.s2post=pnorm(sum((1/(SE^2))*qnorm(1-sp_pval))/sqrt(sum(1/SE^2)), lower.tail=FALSE), fisher=-2*sum(log(sp_pval)), numcontacts=n(), max_B_bin=CID2[which.max(abs(sp_beta))], max_B_FDR=sp_BH_pval[which.max(abs(sp_beta))], max_B=sp_beta[which.max(abs(sp_beta))]) -> cbin.info.VC
full_join(cbin.info.VC, Cstreams.VC, by="CID") -> cbin.full.info.VC
left_join(cgene.hic.overlap.VC, cbin.full.info.VC, by="CID") -> chimpgenes.hic.VC
colnames(chimpgenes.hic.VC)[1:5] <- c("genes", "Hchr", "Hstart", "Hend", "Hstrand") #Fix column names for what was just created
#Now, combine chimpgenes.hic.full and humgenes.hic.full before a final left_join on detable:
full_join(humgenes.VC.full, chimpgenes.hic.VC, by="genes", suffix=c(".H", ".C")) -> genes.hic.VC
full_join(humgenes.KR.full, chimpgenes.hic.KR, by="genes", suffix=c(".H", ".C")) -> genes.hic.KR
###Final join of human and chimp values for both:
left_join(detable, genes.hic.VC, by="genes") -> gene.hic.VC
left_join(detable, genes.hic.KR, by="genes") -> gene.hic.KR
#Clean this dataframe up, removing rows where there is absolutely no Hi-C information for the gene.
filt.VC <- rowSums(is.na(gene.hic.VC)) #51 NA values are found when there is absolutely no Hi-C information.
filt.KR <- rowSums(is.na(gene.hic.KR)) #same.
filt.VC <- which(filt.VC==51)
filt.KR <- which(filt.KR==51)
gene.hic.VC <- gene.hic.VC[-filt.VC,]
gene.hic.KR <- gene.hic.KR[-filt.KR,]
saveRDS(gene.hic.KR, "~/Desktop/Hi-C/gene.hic.filt.KR.RDS")
saveRDS(gene.hic.VC, "~/Desktop/Hi-C/gene.hic.filt.VC.RDS")Differential Expression-Differential Hi-C Enrichment Analyses
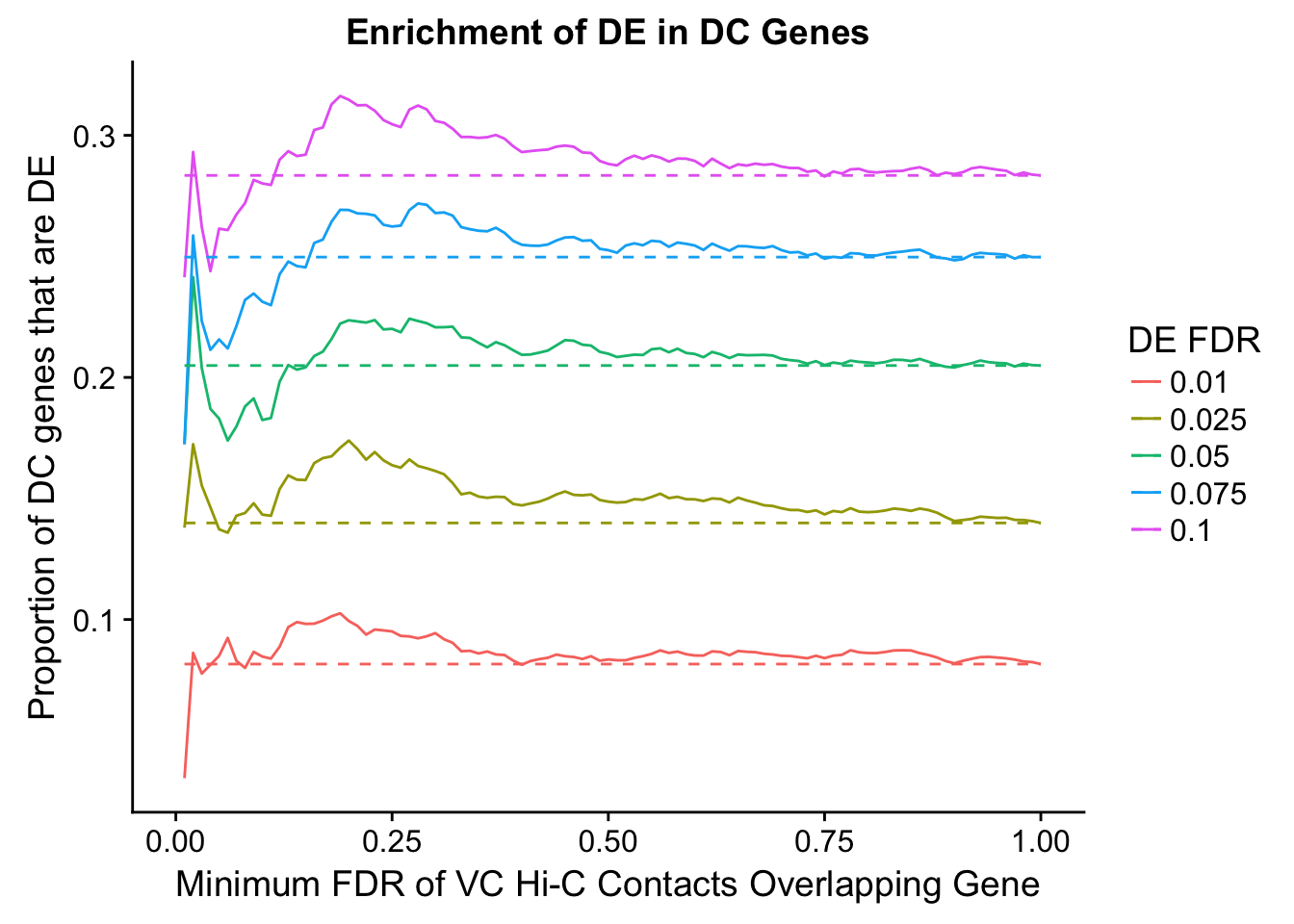
Now I look for enrichment of DHi-C in DE genes using a variety of different metrics to call DHi-C. I now look to see if genes that are differentially expressed are also differential in Hi-C contacts (DHi-C). That is to say, are differentially expressed genes enriched in their overlapping bins for Hi-C contacts that are also differential between the species? To do this I utilize p-values from my prior linear modeling as well as previous RNA-seq analysis. I construct a function to calculate proportions of DE and DHi-C genes, as well as a function to plot this out in a variety of different ways.
####Enrichment analyses!
#A function for calculating proportion of DE genes that are DHi-C under a variety of different paradigms. Accounts for when no genes are DHi-C and when all genes are DHi-C. Returns the proportion of DE genes that are also DHi-C, as well as the expected proportion based on conditional probability alone.
prop.calculator <- function(de.vec, hic.vec, i, k){
my.result <- data.frame(prop=NA, exp.prop=NA, chisq.p=NA, Dneither=NA, DE=NA, DHiC=NA, Dboth=NA)
bad.indices <- which(is.na(hic.vec)) #First obtain indices where Hi-C info is missing, if there are any, then remove from both vectors.
if(length(bad.indices>0)){
de.vec <- de.vec[-bad.indices]
hic.vec <- hic.vec[-bad.indices]}
de.vec <- ifelse(de.vec<=i, 1, 0)
hic.vec <- ifelse(hic.vec<=k, 1, 0)
if(sum(hic.vec, na.rm=TRUE)==0){#The case where no genes show up as DHi-C.
my.result[1,] <- c(0, 0, 0, sum(de.vec==0, na.rm=TRUE), sum(de.vec==1, na.rm=TRUE), 0, 0) #Since no genes are DHi-C, the proportion is 0 and our expectation is 0, set p-val=0 since it's irrelevant.
}
else if(sum(hic.vec)==length(hic.vec)){ #The case where every gene shows up as DHi-C
my.result[1,] <- c(1, 1, 0, 0, 0, sum(hic.vec==1&de.vec==0, na.rm = TRUE), sum(de.vec==1&hic.vec==1, na.rm=TRUE)) #If every gene is DHi-C, the observed proportion of DE genes DHi-C is 1, and the expected proportion of DE genes also DHi-C would also be 1 (all DE genes are DHi-C, since all genes are). Again set p-val to 0 since irrelevant comparison.
}
else{#The typical case, where we get an actual table
mytable <- table(as.data.frame(cbind(de.vec, hic.vec)))
my.result[1,1] <- mytable[2,2]/sum(mytable[2,]) #The observed proportion of DE genes that are also DHi-C. # that are both/total # DE
my.result[1,2] <- (((sum(mytable[2,])/sum(mytable))*((sum(mytable[,2])/sum(mytable))))*sum(mytable))/sum(mytable[2,]) #The expected proportion: (p(DE) * p(DHiC)) * total # genes / # DE genes
my.result[1,3] <- chisq.test(mytable)$p.value
my.result[1,4] <- mytable[1,1]
my.result[1,5] <- mytable[2,1]
my.result[1,6] <- mytable[1,2]
my.result[1,7] <- mytable[2,2]
}
return(my.result)
}
#This is a function that computes observed and expected proportions of DE and DHiC enrichments, and spits out a variety of different visualizations for them. As input it takes a dataframe, the names of its DHiC and DE p-value columns, and a name to represent the type of Hi-C contact summary for the gene that ends up on the x-axis of all the plots.
enrichment.plotter <- function(df, HiC_col, DE_col, xlab, xmax=0.3, i=c(0.01, 0.025, 0.05, 0.075, 0.1), k=seq(0.01, 1, 0.01)){
enrich.table <- data.frame(DEFDR = c(rep(i[1], 100), rep(i[2], 100), rep(i[3], 100), rep(i[4], 100), rep(i[5], 100)), DHICFDR=rep(k, 5), prop.obs=NA, prop.exp=NA, chisq.p=NA, Dneither=NA, DE=NA, DHiC=NA, Dboth=NA)
for(de.FDR in i){
for(hic.FDR in k){
enrich.table[which(enrich.table$DEFDR==de.FDR&enrich.table$DHICFDR==hic.FDR), 3:9] <- prop.calculator(df[,DE_col], df[,HiC_col], de.FDR, hic.FDR)
}
}
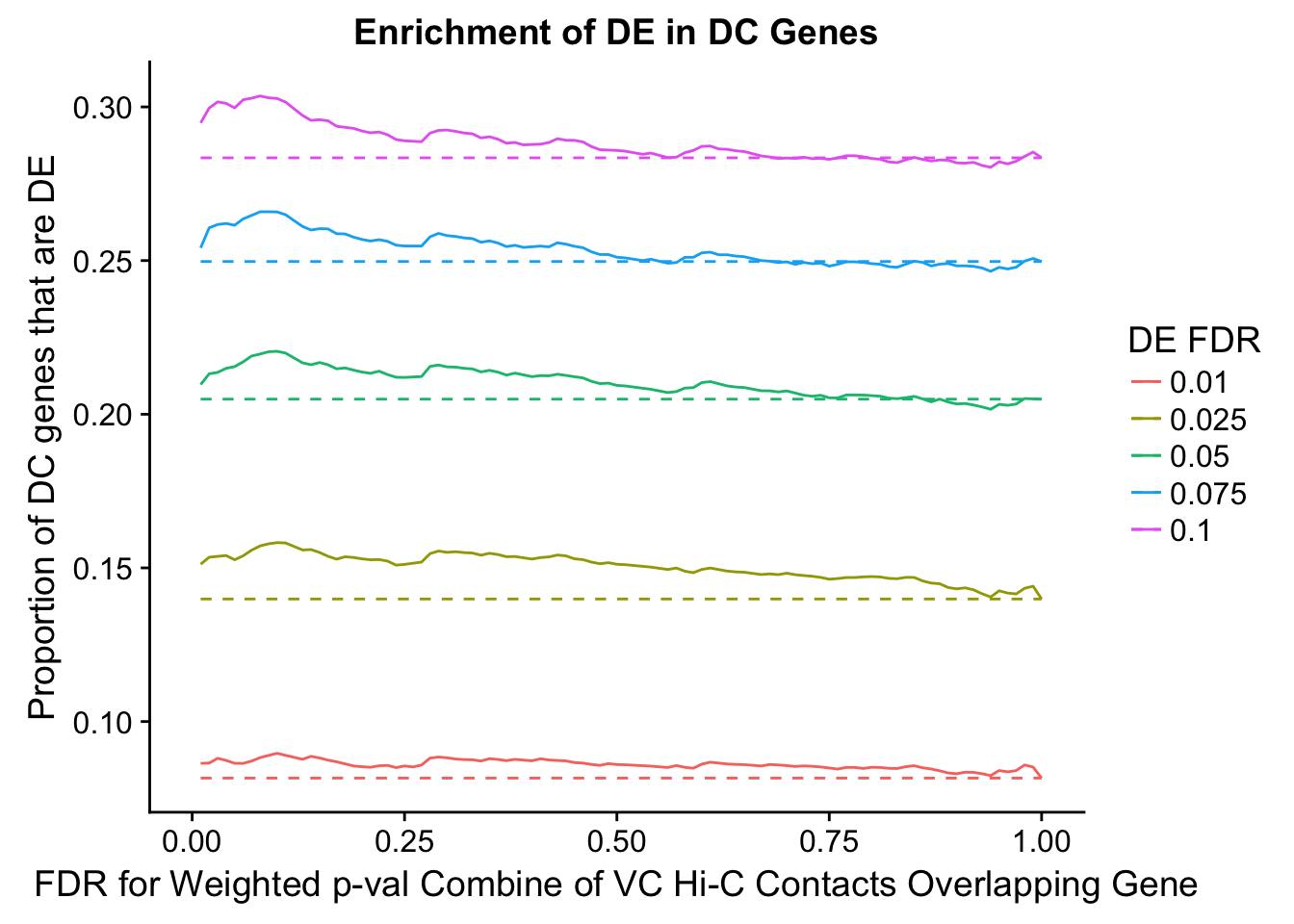
des.enriched <- ggplot(data=enrich.table, aes(x=DHICFDR, y=prop.obs, group=as.factor(DEFDR), color=as.factor(DEFDR))) +geom_line()+ geom_line(aes(y=prop.exp), linetype="dashed", size=0.5) + ggtitle("Enrichment of DC in DE Genes") + xlab(xlab) + ylab("Proportion of DE genes that are DC") + guides(color=guide_legend(title="FDR for DE Genes"))
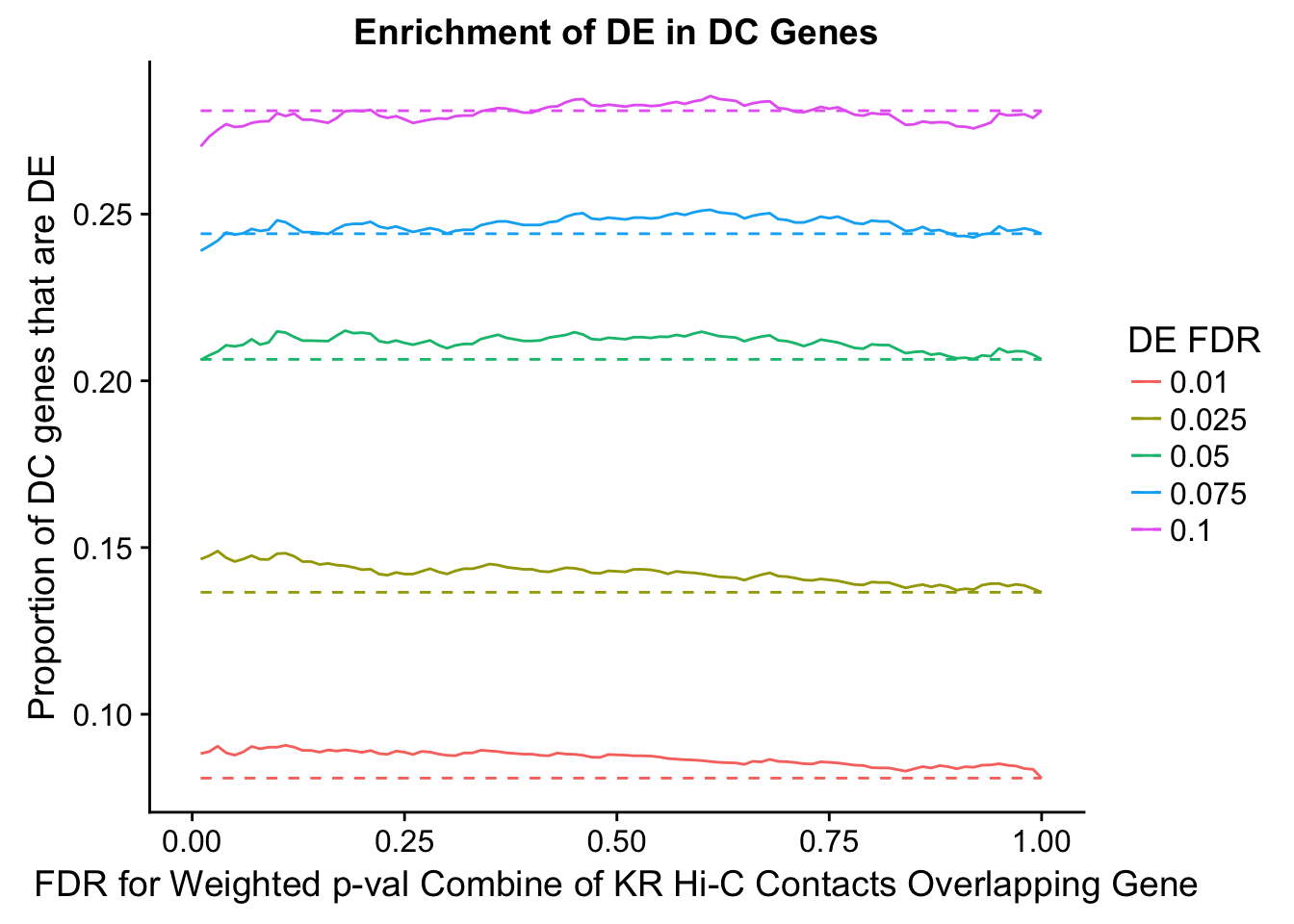
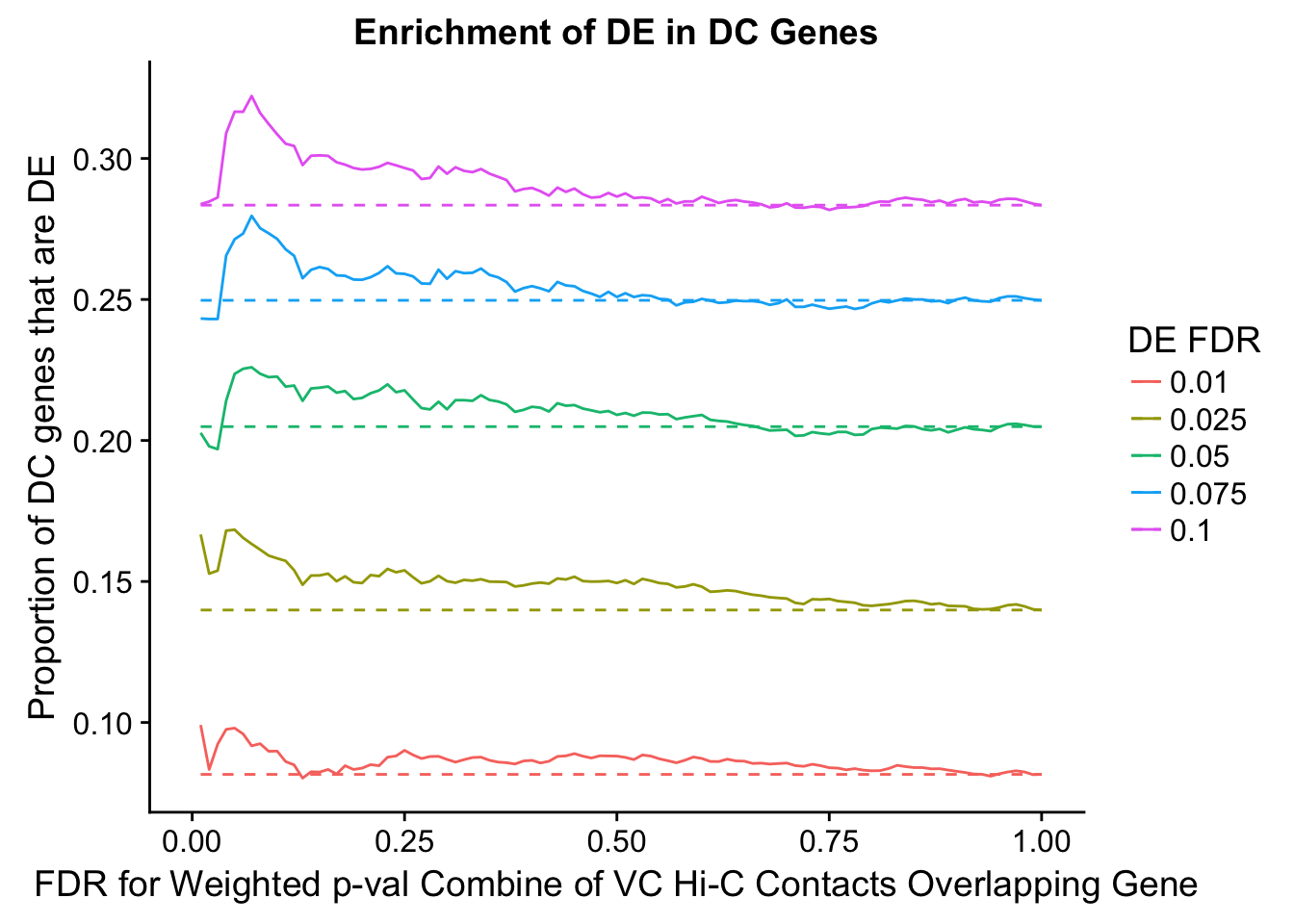
dhics.enriched <- ggplot(data=enrich.table, aes(x=DHICFDR, y=Dboth/(Dboth+DHiC), group=as.factor(DEFDR), color=as.factor(DEFDR))) + geom_line() + geom_line(aes(y=(((((DE+Dboth)/(Dneither+DE+DHiC+Dboth))*((DHiC+Dboth)/(Dneither+DE+DHiC+Dboth)))*(Dneither+DE+DHiC+Dboth))/(DHiC+Dboth))), linetype="dashed") + ylab("Proportion of DC genes that are DE") +xlab(xlab) + ggtitle("Enrichment of DE in DC Genes") + coord_cartesian(xlim=c(0, xmax)) + guides(color=guide_legend(title="DE FDR"))
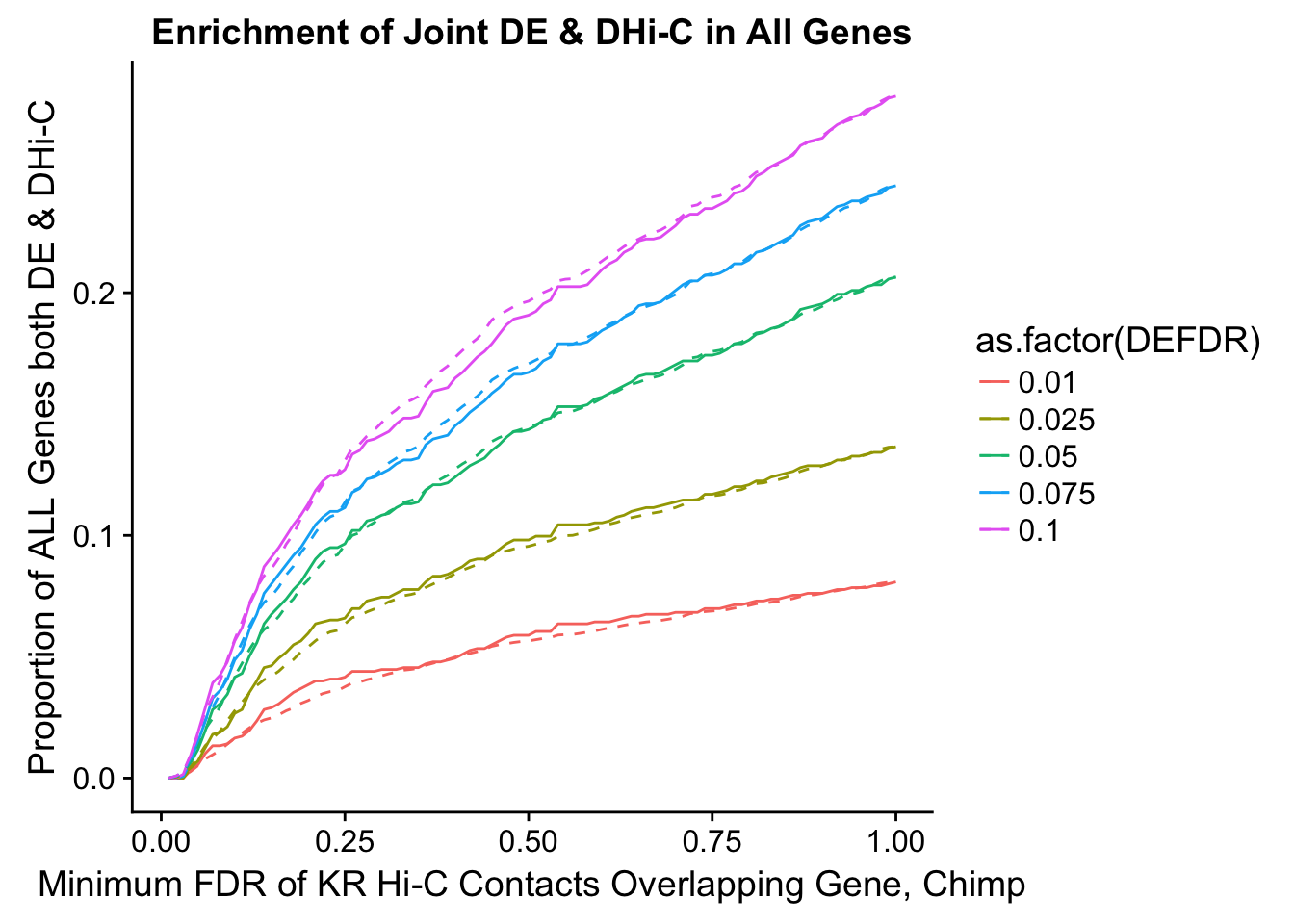
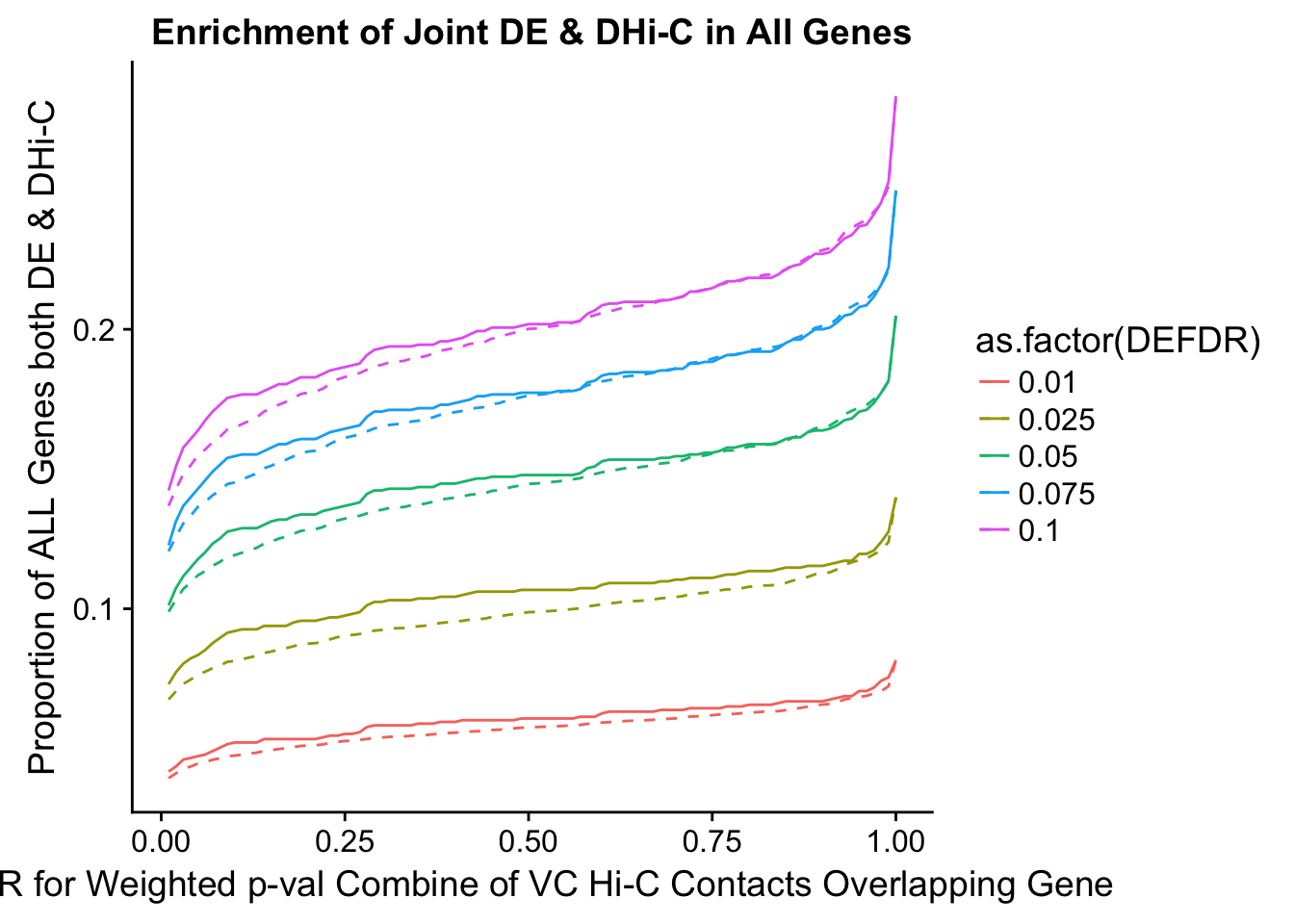
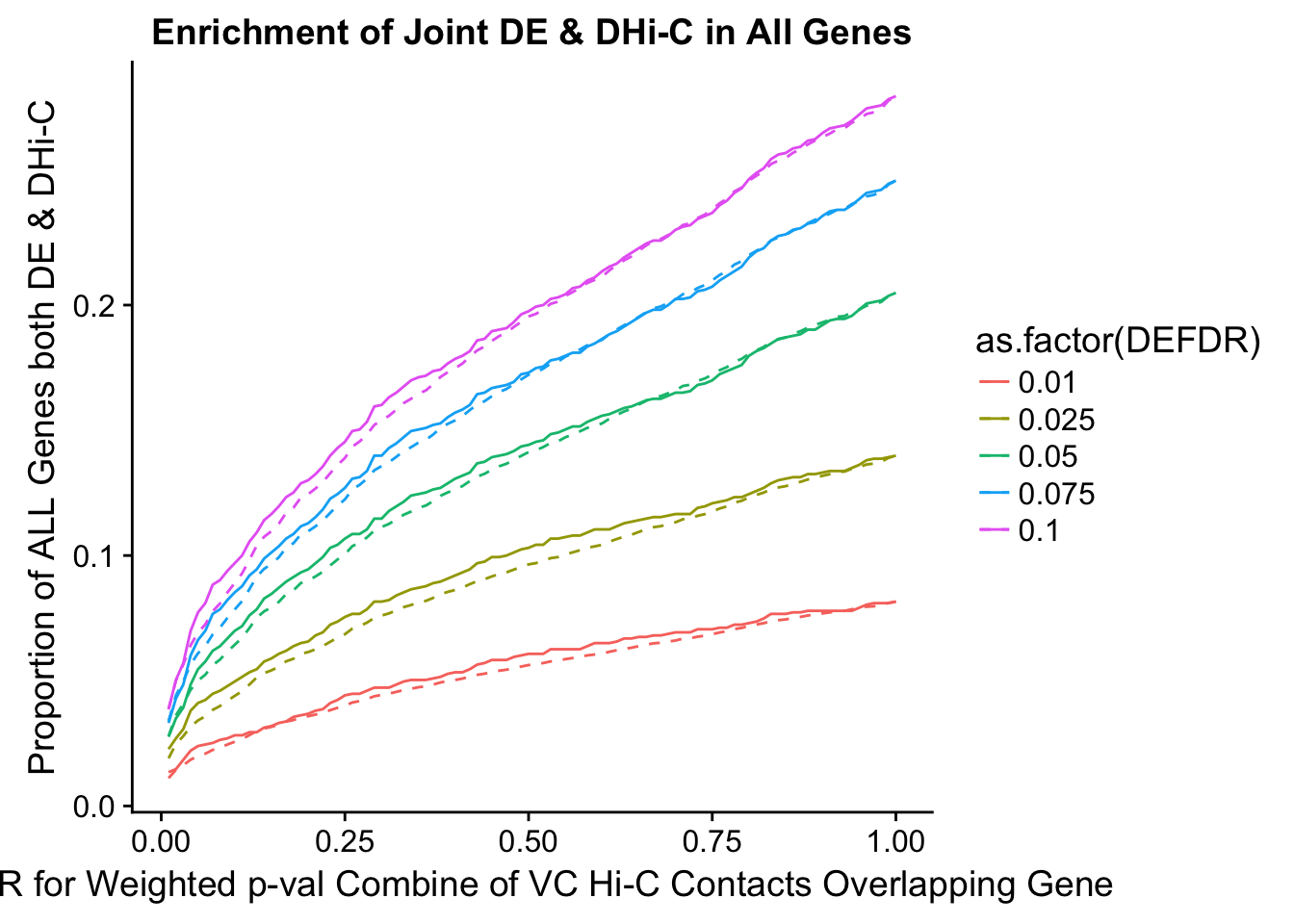
joint.enriched <- ggplot(data=enrich.table, aes(x=DHICFDR, y=Dboth/(Dneither+DE+DHiC+Dboth), group=as.factor(DEFDR), color=as.factor(DEFDR))) + geom_line() + ylab("Proportion of ALL Genes both DE & DHi-C") + xlab(xlab) + geom_line(aes(y=((DE+Dboth)/(Dneither+DE+DHiC+Dboth))*((DHiC+Dboth)/(Dneither+DE+DHiC+Dboth))), linetype="dashed") + ggtitle("Enrichment of Joint DE & DHi-C in All Genes")
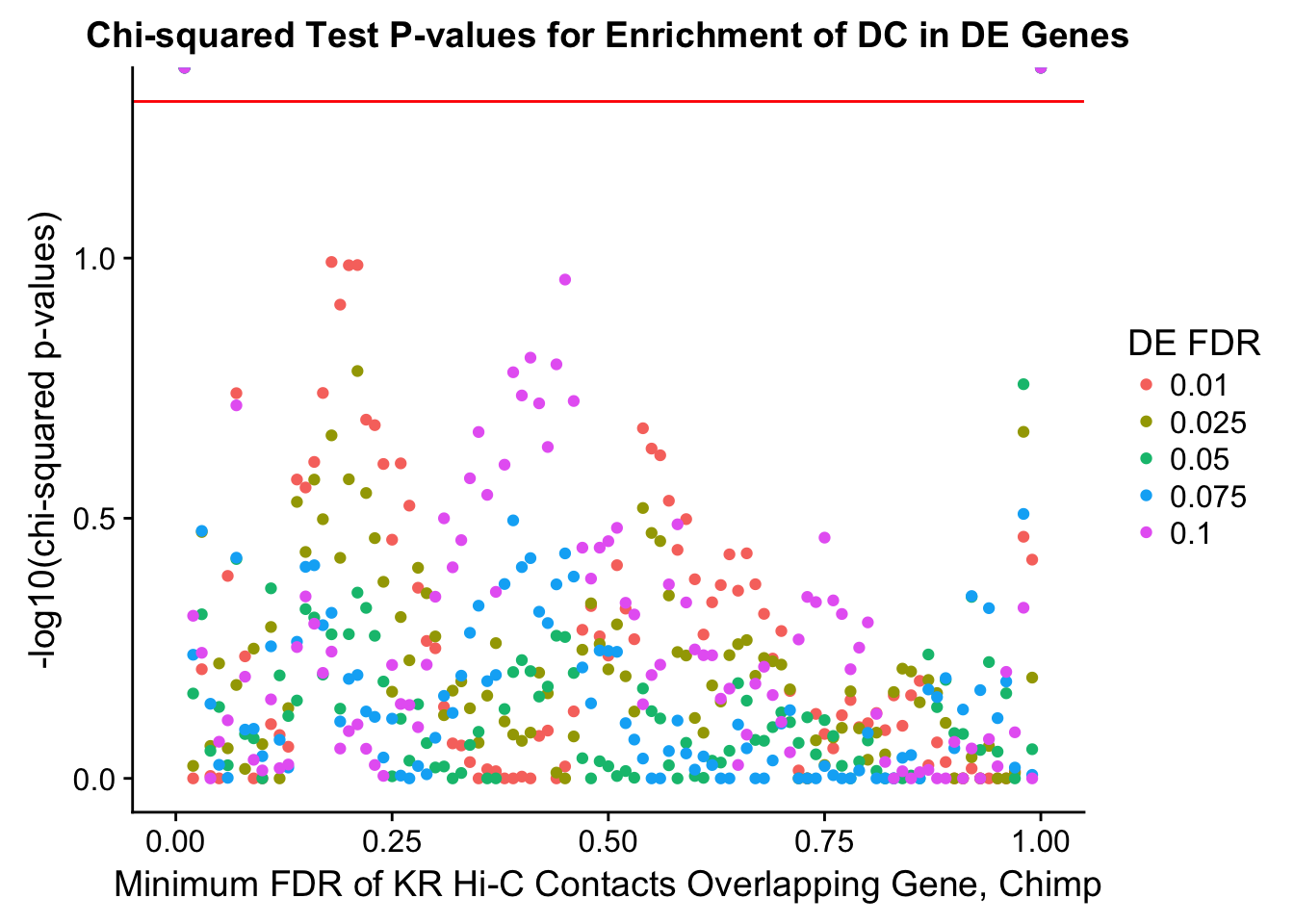
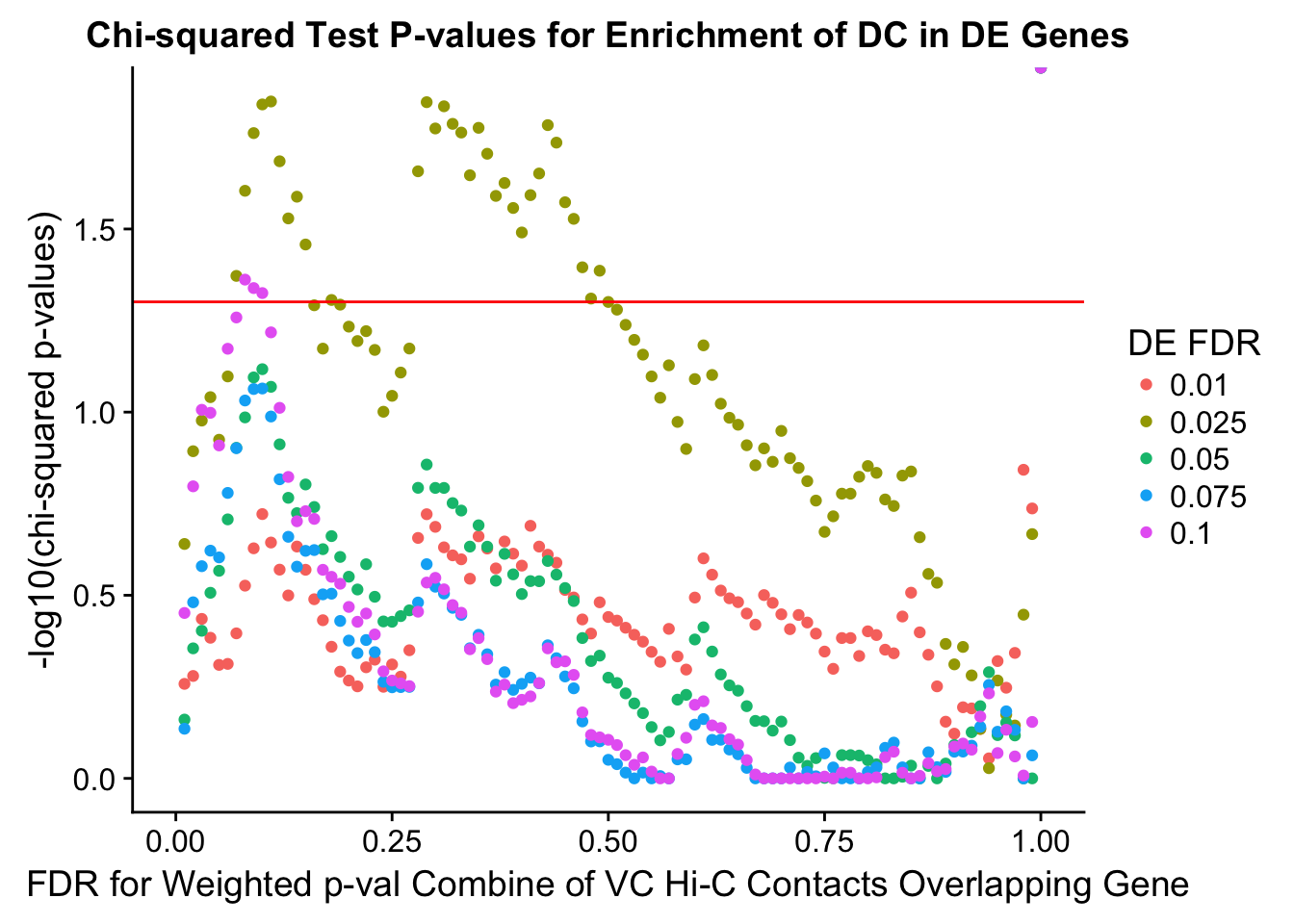
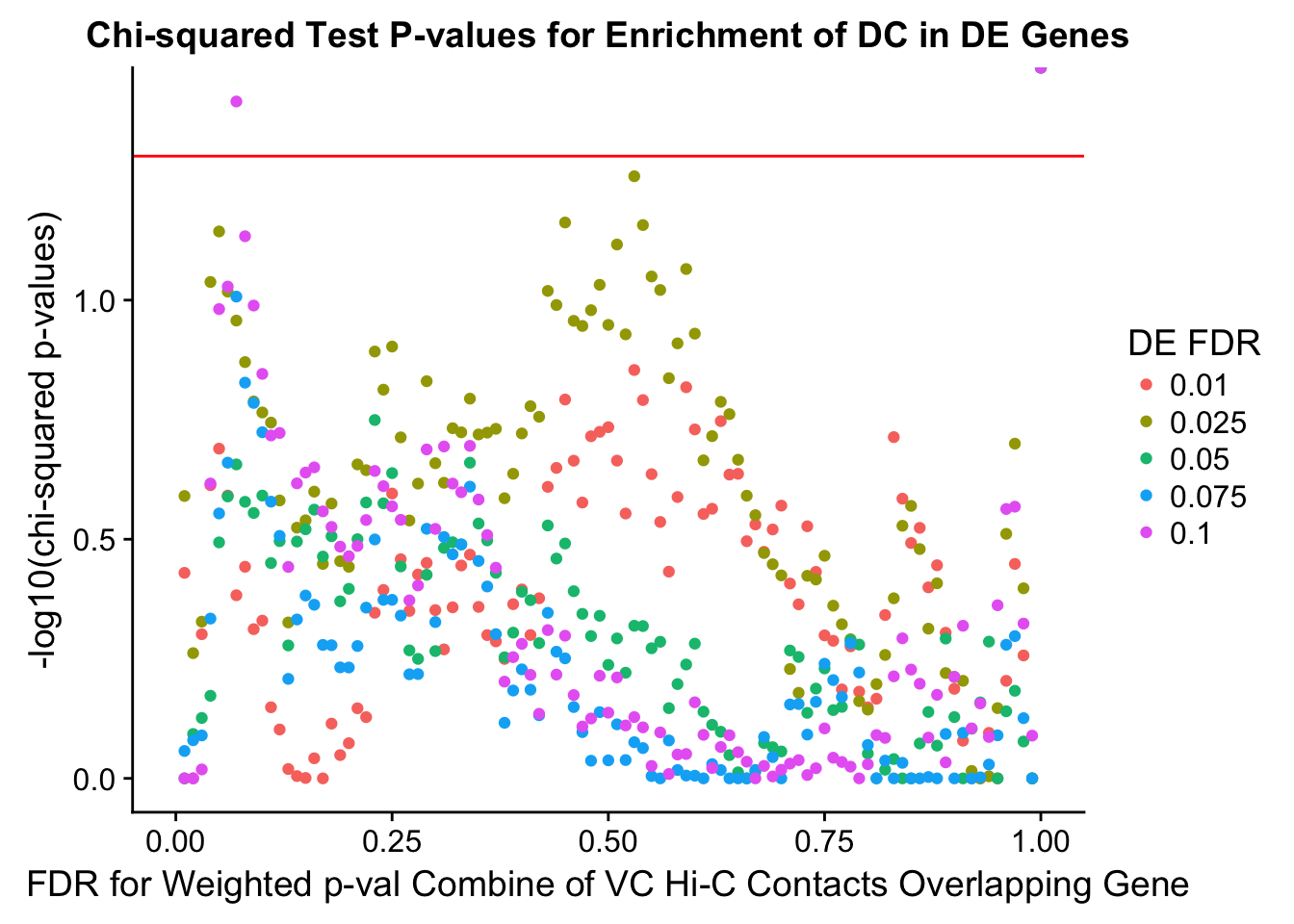
chisq.p <- ggplot(data=enrich.table, aes(x=DHICFDR, y=-log10(chisq.p), group=as.factor(DEFDR), color=as.factor(DEFDR))) + geom_point() + geom_hline(yintercept=-log10(0.05), color="red") + ggtitle("Chi-squared Test P-values for Enrichment of DC in DE Genes") + xlab(xlab) + ylab("-log10(chi-squared p-values)") + coord_cartesian(xlim=c(0, xmax)) + guides(color=guide_legend(title="DE FDR"))
print(des.enriched)
print(dhics.enriched)
print(joint.enriched)
print(chisq.p)
print(enrich.table[which(enrich.table$DEFDR==0.025),]) #Added to figure out comparison for the paper.
}
#Visualization of enrichment of DE/DC in one another. For most of these, using the gene.hic.filt df is sufficient as their Hi-C FDR numbers are the same. For the upstream genes it's a little more complicated because gene.hic.filt doesn't incorporate strand information on the genes, so use the specific US dfs for that, with the USFDR column.
enrichment.plotter(gene.hic.VC, "min_FDR.H", "adj.P.Val", "Minimum FDR of VC Hi-C Contacts Overlapping Gene", xmax=1)Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect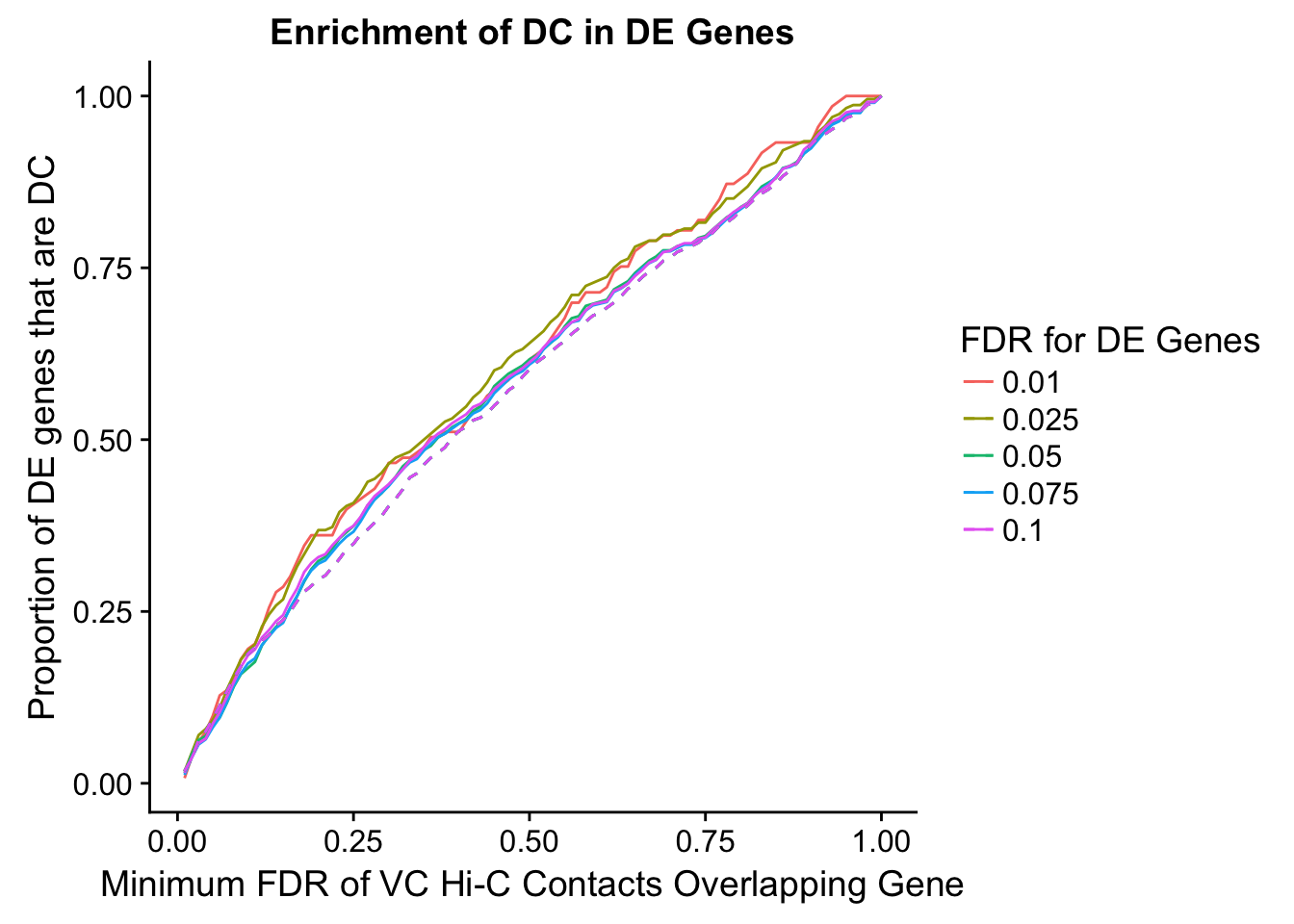
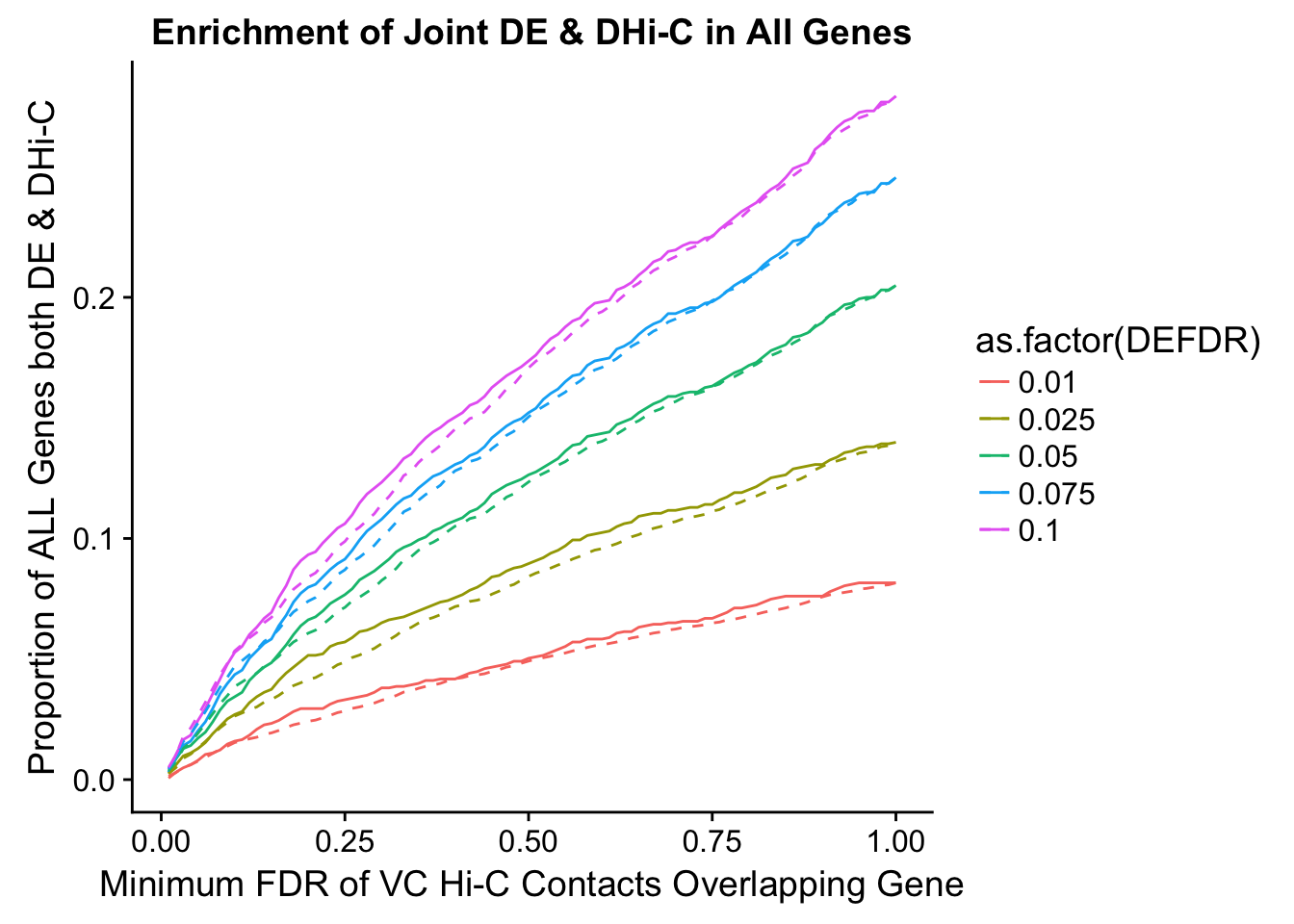
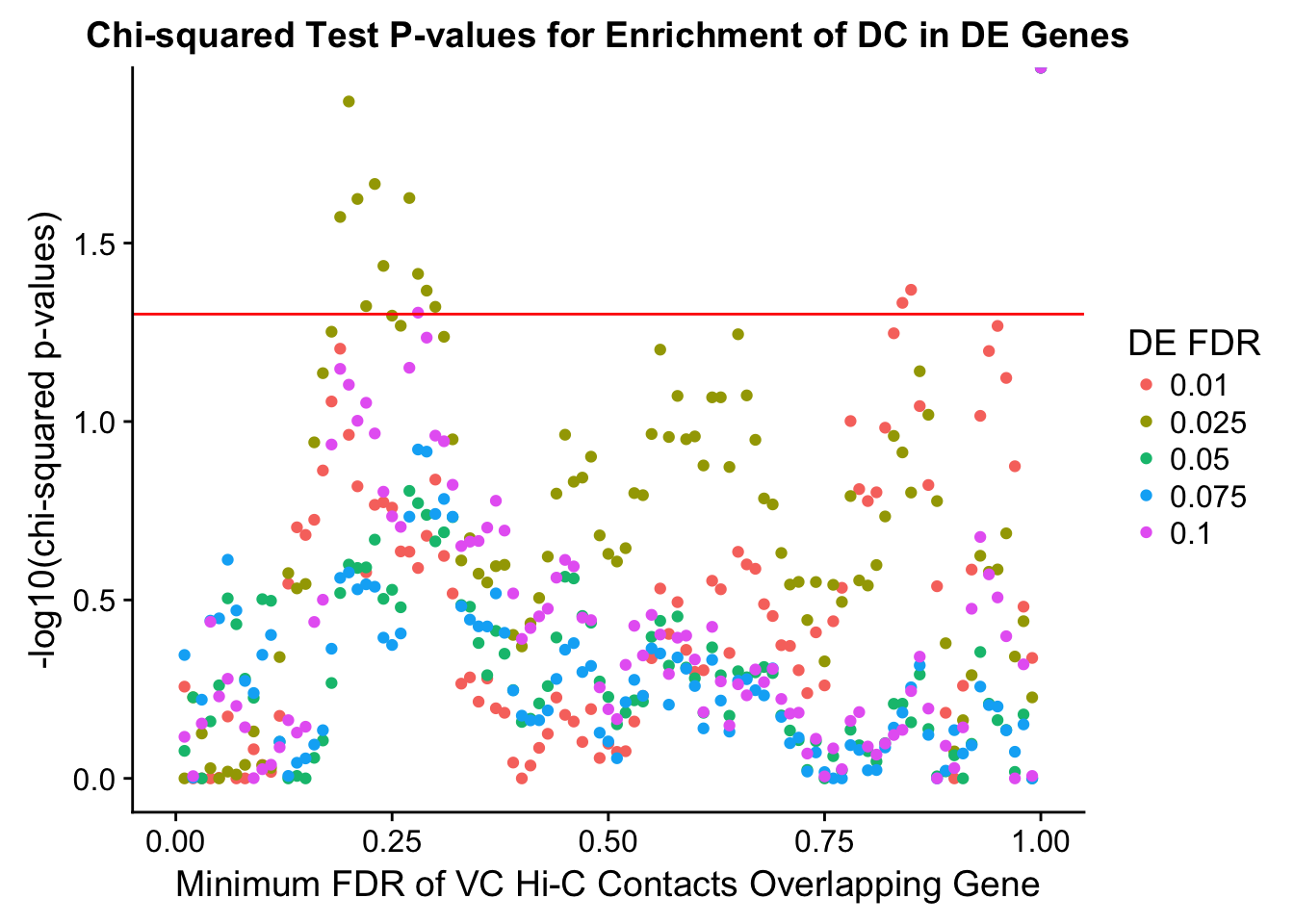
DEFDR DHICFDR prop.obs prop.exp chisq.p Dneither DE DHiC Dboth
101 0.025 0.01 0.01754386 0.01779141 1.00000000 1377 224 25 4
102 0.025 0.02 0.04385965 0.03558282 0.59285521 1354 218 48 10
103 0.025 0.03 0.07017544 0.06319018 0.74844939 1315 212 87 16
104 0.025 0.04 0.07894737 0.07546012 0.93641307 1297 210 105 18
105 0.025 0.05 0.09210526 0.09386503 1.00000000 1270 207 132 21
106 0.025 0.06 0.10964912 0.11288344 0.95727298 1243 203 159 25
107 0.025 0.07 0.13596491 0.13312883 0.97541215 1216 197 186 31
108 0.025 0.08 0.15789474 0.15337423 0.91624724 1188 192 214 36
109 0.025 0.09 0.17982456 0.16993865 0.73876738 1166 187 236 41
110 0.025 0.10 0.19298246 0.18834356 0.91887460 1139 184 263 44
111 0.025 0.11 0.20175439 0.19754601 0.93431729 1126 182 276 46
112 0.025 0.12 0.22807018 0.20736196 0.45714447 1116 176 286 52
113 0.025 0.13 0.24561404 0.21533742 0.26599015 1107 172 295 56
114 0.025 0.14 0.25877193 0.22944785 0.29347111 1087 169 315 59
115 0.025 0.15 0.26754386 0.23742331 0.28524810 1076 167 326 61
116 0.025 0.16 0.29385965 0.24969325 0.11437228 1062 161 340 67
117 0.025 0.17 0.31578947 0.26503067 0.07320091 1042 156 360 72
118 0.025 0.18 0.33333333 0.27852761 0.05601957 1024 152 378 76
119 0.025 0.19 0.35087719 0.28711656 0.02671540 1014 148 388 80
120 0.025 0.20 0.36842105 0.29631902 0.01268109 1003 144 399 84
121 0.025 0.21 0.36842105 0.30245399 0.02378674 993 144 409 84
122 0.025 0.22 0.37280702 0.31411043 0.04748361 975 143 427 85
123 0.025 0.23 0.39473684 0.32638037 0.02159607 960 138 442 90
124 0.025 0.24 0.40350877 0.34049080 0.03663515 939 136 463 92
125 0.025 0.25 0.40789474 0.34846626 0.05049965 927 135 475 93
126 0.025 0.26 0.42105263 0.36196319 0.05390464 908 132 494 96
127 0.025 0.27 0.43859649 0.36932515 0.02364280 900 128 502 100
128 0.025 0.28 0.44298246 0.37914110 0.03856780 885 127 517 101
129 0.025 0.29 0.45175439 0.38895706 0.04297312 871 125 531 103
130 0.025 0.30 0.46491228 0.40306748 0.04770651 851 122 551 106
131 0.025 0.31 0.47368421 0.41411043 0.05787378 835 120 567 108
132 0.025 0.32 0.47807018 0.42760736 0.11216822 814 119 588 109
133 0.025 0.33 0.48245614 0.44478528 0.24509083 787 118 615 110
134 0.025 0.34 0.49122807 0.45092025 0.21234769 779 116 623 112
135 0.025 0.35 0.50000000 0.46380368 0.26693651 760 114 642 114
136 0.025 0.36 0.50877193 0.47361963 0.28249414 746 112 656 116
137 0.025 0.37 0.51754386 0.48036810 0.25428445 737 110 665 118
138 0.025 0.38 0.52631579 0.48895706 0.25205891 725 108 677 120
139 0.025 0.39 0.53070175 0.50245399 0.39620517 704 107 698 121
140 0.025 0.40 0.53947368 0.51288344 0.42678869 689 105 713 123
141 0.025 0.41 0.54824561 0.51840491 0.36764763 682 103 720 125
142 0.025 0.42 0.56140351 0.52822086 0.31215477 669 100 733 128
143 0.025 0.43 0.57017544 0.53190184 0.23908682 665 98 737 130
144 0.025 0.44 0.58333333 0.53803681 0.15923876 658 95 744 133
145 0.025 0.45 0.60087719 0.54969325 0.10888967 643 91 759 137
146 0.025 0.46 0.60526316 0.55889571 0.14747350 629 90 773 138
147 0.025 0.47 0.61842105 0.57177914 0.14360002 611 87 791 141
148 0.025 0.48 0.62719298 0.57852761 0.12545552 602 85 800 143
149 0.025 0.49 0.63157895 0.59141104 0.20848034 582 84 820 144
150 0.025 0.50 0.64035088 0.60245399 0.23490791 566 82 836 146
151 0.025 0.51 0.64912281 0.61226994 0.24678832 552 80 850 148
152 0.025 0.52 0.65789474 0.61963190 0.22641009 542 78 860 150
153 0.025 0.53 0.67105263 0.62699387 0.15869530 533 75 869 153
154 0.025 0.54 0.67982456 0.63619632 0.16083799 520 73 882 155
155 0.025 0.55 0.69298246 0.64355828 0.10836950 511 70 891 158
156 0.025 0.56 0.71052632 0.65398773 0.06288059 498 66 904 162
157 0.025 0.57 0.71052632 0.66196319 0.11049384 485 66 917 162
158 0.025 0.58 0.72368421 0.67177914 0.08476833 472 63 930 165
159 0.025 0.59 0.72807018 0.68036810 0.11208973 459 62 943 166
160 0.025 0.60 0.73245614 0.68466258 0.11008002 453 61 949 167
161 0.025 0.61 0.73684211 0.69202454 0.13277696 442 60 960 168
162 0.025 0.62 0.75000000 0.69938650 0.08555825 433 57 969 171
163 0.025 0.63 0.75877193 0.70858896 0.08553381 420 55 982 173
164 0.025 0.64 0.76315789 0.71963190 0.13408581 403 54 999 174
165 0.025 0.65 0.78070175 0.72638037 0.05694501 396 50 1006 178
166 0.025 0.66 0.78508772 0.73619632 0.08448235 381 49 1021 179
167 0.025 0.67 0.78947368 0.74478528 0.11252583 368 48 1034 180
168 0.025 0.68 0.78947368 0.75030675 0.16429036 359 48 1043 180
169 0.025 0.69 0.79824561 0.76012270 0.17070008 345 46 1057 182
170 0.025 0.70 0.79824561 0.76503067 0.23354632 337 46 1065 182
171 0.025 0.71 0.80263158 0.77300613 0.28631683 325 45 1077 183
172 0.025 0.72 0.80701754 0.77730061 0.28144899 319 44 1083 184
173 0.025 0.73 0.80701754 0.78159509 0.35998888 312 44 1090 184
174 0.025 0.74 0.81578947 0.78650307 0.28171362 306 42 1096 186
175 0.025 0.75 0.81578947 0.79570552 0.47000885 291 42 1111 186
176 0.025 0.76 0.82894737 0.80061350 0.28676648 286 39 1116 189
177 0.025 0.77 0.83771930 0.81165644 0.32023462 270 37 1132 191
178 0.025 0.78 0.85087719 0.81533742 0.16174984 267 34 1135 194
179 0.025 0.79 0.85087719 0.82331288 0.27878955 254 34 1148 194
180 0.025 0.80 0.85964912 0.83312883 0.28811359 240 32 1162 196
181 0.025 0.81 0.86842105 0.84049080 0.25244409 230 30 1172 198
182 0.025 0.82 0.88157895 0.85030675 0.18449850 217 27 1185 201
183 0.025 0.83 0.89473684 0.85828221 0.10972262 207 24 1195 204
184 0.025 0.84 0.89912281 0.86441718 0.12204781 198 23 1204 205
185 0.025 0.85 0.90350877 0.87239264 0.15813738 186 22 1216 206
186 0.025 0.86 0.92105263 0.88343558 0.07229154 172 18 1230 210
187 0.025 0.87 0.92543860 0.89141104 0.09572916 160 17 1242 211
188 0.025 0.88 0.92982456 0.90245399 0.16709282 143 16 1259 212
189 0.025 0.89 0.93421053 0.91840491 0.41815957 118 15 1284 213
190 0.025 0.90 0.93421053 0.92883436 0.84024590 101 15 1301 213
191 0.025 0.91 0.94736842 0.93926380 0.68696721 87 12 1315 216
192 0.025 0.92 0.95614035 0.94478528 0.51368387 80 10 1322 218
193 0.025 0.93 0.96929825 0.95153374 0.23777935 72 7 1330 221
194 0.025 0.94 0.97368421 0.95766871 0.26368339 63 6 1339 222
195 0.025 0.95 0.98245614 0.96809816 0.25973393 48 4 1354 224
196 0.025 0.96 0.98684211 0.97177914 0.20576095 43 3 1359 225
197 0.025 0.97 0.98684211 0.97791411 0.45558545 33 3 1369 225
198 0.025 0.98 0.99561404 0.98711656 0.36271687 20 1 1382 227
199 0.025 0.99 0.99561404 0.99018405 0.59294582 15 1 1387 227
200 0.025 1.00 1.00000000 1.00000000 0.00000000 0 0 1402 228enrichment.plotter(gene.hic.KR, "min_FDR.H", "adj.P.Val", "Minimum FDR of KR Hi-C Contacts Overlapping Gene, Chimp", xmax=1)Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect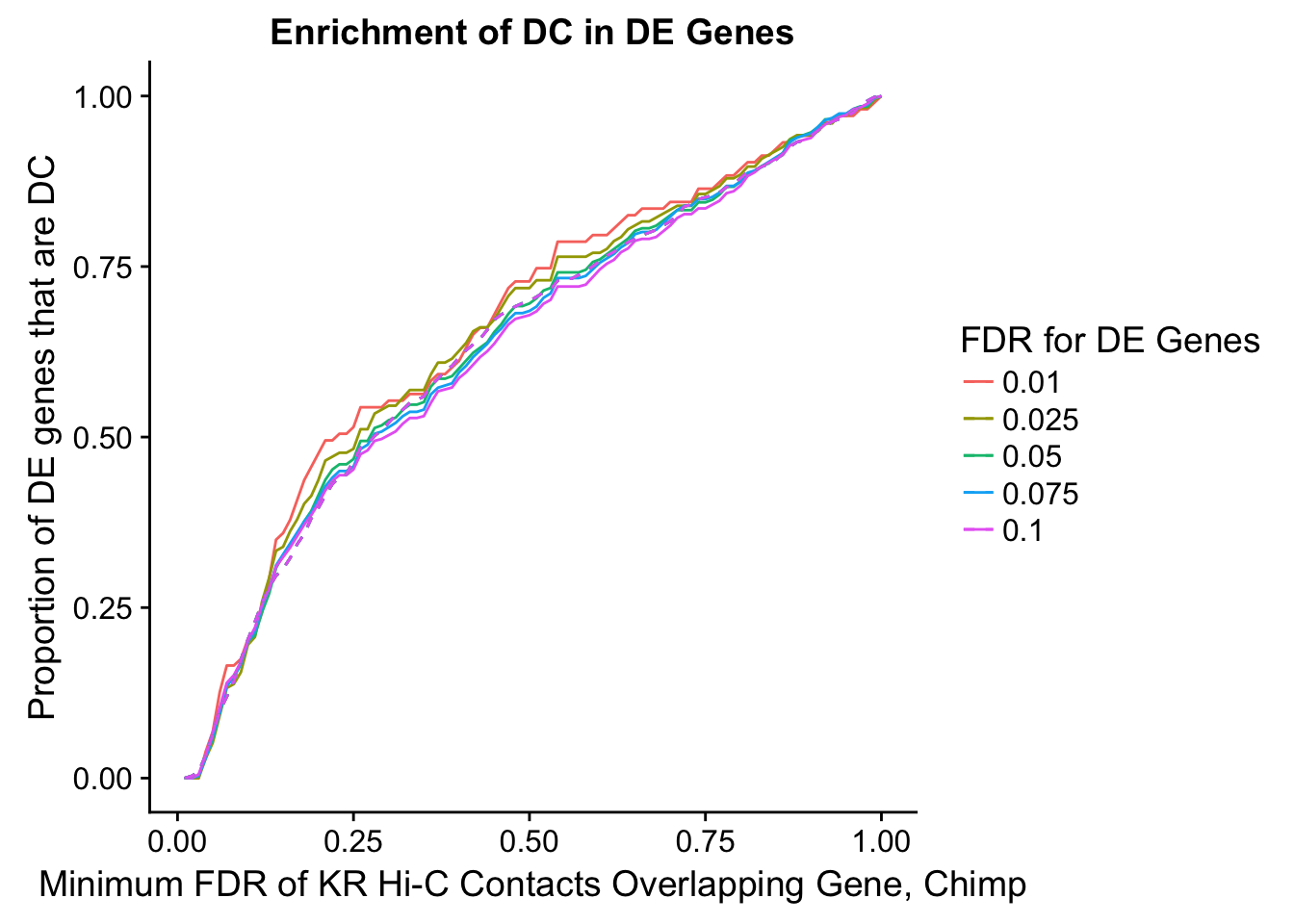
Warning: Removed 5 rows containing missing values (geom_path).Warning: Removed 5 rows containing missing values (geom_path).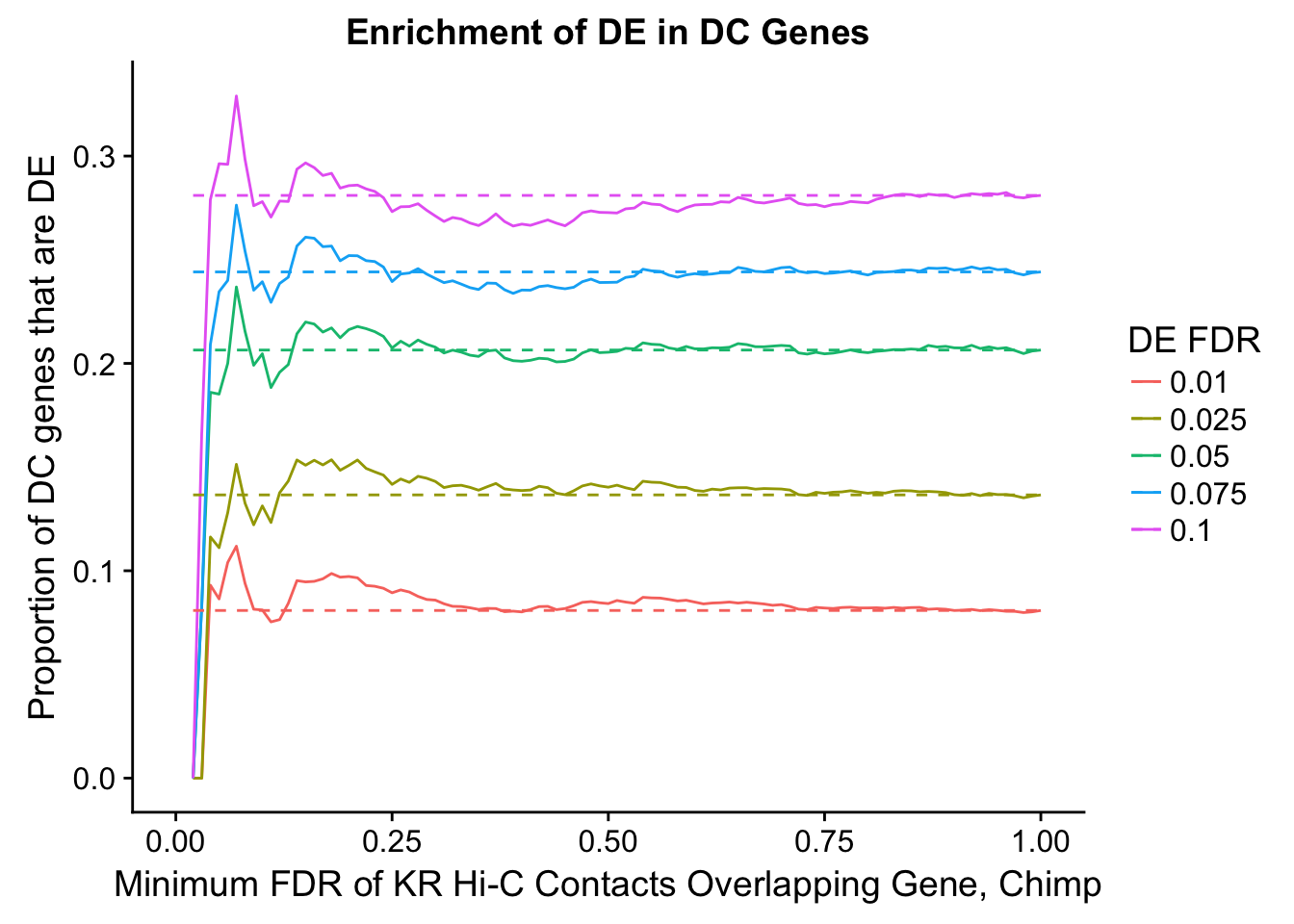
DEFDR DHICFDR prop.obs prop.exp chisq.p Dneither DE DHiC Dboth
101 0.025 0.01 0.00000000 0.000000000 0.0000000 1100 174 0 0
102 0.025 0.02 0.00000000 0.003139717 0.9461552 1096 174 4 0
103 0.025 0.03 0.00000000 0.009419152 0.3360654 1088 174 12 0
104 0.025 0.04 0.02873563 0.033751962 0.8662375 1062 169 38 5
105 0.025 0.05 0.05172414 0.063579278 0.6012921 1028 165 72 9
106 0.025 0.06 0.09195402 0.098116170 0.8752942 991 158 109 16
107 0.025 0.07 0.13218391 0.119309262 0.6613957 971 151 129 23
108 0.025 0.08 0.13793103 0.142072214 0.9588927 943 150 157 24
109 0.025 0.09 0.15517241 0.173469388 0.5631071 906 147 194 27
110 0.025 0.10 0.19540230 0.203296703 0.8594275 875 140 225 34
111 0.025 0.11 0.20689655 0.229199372 0.5116904 844 138 256 36
112 0.025 0.12 0.25862069 0.256671900 1.0000000 818 129 282 45
113 0.025 0.13 0.29310345 0.279434851 0.7327157 795 123 305 51
114 0.025 0.14 0.33333333 0.296703297 0.2941624 780 116 320 58
115 0.025 0.15 0.33908046 0.306907378 0.3671477 768 115 332 59
116 0.025 0.16 0.36206897 0.322605965 0.2665157 752 111 348 63
117 0.025 0.17 0.37931034 0.343014129 0.3175669 729 108 371 66
118 0.025 0.18 0.40229885 0.357927786 0.2191319 714 104 386 70
119 0.025 0.19 0.41379310 0.380690738 0.3768152 687 102 413 72
120 0.025 0.20 0.43678161 0.395604396 0.2661303 672 98 428 76
121 0.025 0.21 0.46551724 0.414442700 0.1648331 653 93 447 81
122 0.025 0.22 0.47126437 0.430926217 0.2828294 633 92 467 82
123 0.025 0.23 0.47701149 0.441130298 0.3453178 621 91 479 83
124 0.025 0.24 0.47701149 0.445839874 0.4189822 615 91 485 83
125 0.025 0.25 0.48275862 0.465463108 0.6814792 591 90 509 84
126 0.025 0.26 0.51149425 0.484301413 0.4896858 572 85 528 89
127 0.025 0.27 0.51149425 0.489795918 0.5929399 565 85 535 89
128 0.025 0.28 0.53448276 0.501569859 0.3937285 554 81 546 93
129 0.025 0.29 0.54022989 0.510204082 0.4406704 544 80 556 94
130 0.025 0.30 0.54597701 0.521193093 0.5335250 531 79 569 95
131 0.025 0.31 0.54597701 0.532182104 0.7560128 517 79 583 95
132 0.025 0.32 0.55747126 0.540031397 0.6782184 509 77 591 97
133 0.025 0.33 0.56896552 0.550235479 0.6509207 498 75 602 99
134 0.025 0.34 0.56896552 0.554160126 0.7332758 493 75 607 99
135 0.025 0.35 0.56896552 0.559654631 0.8539491 486 75 614 99
136 0.025 0.36 0.59195402 0.575353218 0.6934009 470 71 630 103
137 0.025 0.37 0.60919540 0.585557300 0.5495944 460 68 640 106
138 0.025 0.38 0.60919540 0.596546311 0.7772778 446 68 654 106
139 0.025 0.39 0.61494253 0.604395604 0.8237142 437 67 663 107
140 0.025 0.40 0.62643678 0.616954474 0.8469684 423 65 677 109
141 0.025 0.41 0.63793103 0.627158556 0.8166244 412 63 688 111
142 0.025 0.42 0.65517241 0.635792779 0.6263015 404 60 696 114
143 0.025 0.43 0.66091954 0.644427002 0.6862983 394 59 706 115
144 0.025 0.44 0.66091954 0.656985871 0.9747102 378 59 722 115
145 0.025 0.45 0.67241379 0.671899529 1.0000000 361 57 739 117
146 0.025 0.46 0.68965517 0.679748823 0.8305636 354 54 746 120
147 0.025 0.47 0.70689655 0.685243328 0.5659419 350 51 750 123
148 0.025 0.48 0.71839080 0.691522763 0.4608209 344 49 756 125
149 0.025 0.49 0.71839080 0.696232339 0.5516460 338 49 762 125
150 0.025 0.50 0.71839080 0.699372057 0.6171827 334 49 766 125
151 0.025 0.51 0.72988506 0.705651491 0.5058383 328 47 772 127
152 0.025 0.52 0.72988506 0.711930926 0.6364056 320 47 780 127
153 0.025 0.53 0.72988506 0.716640502 0.7438865 314 47 786 127
154 0.025 0.54 0.76436782 0.729199372 0.3022179 304 41 796 133
155 0.025 0.55 0.76436782 0.731554160 0.3375076 301 41 799 133
156 0.025 0.56 0.76436782 0.732339089 0.3498790 300 41 800 133
157 0.025 0.57 0.76436782 0.737833595 0.4450468 293 41 807 133
158 0.025 0.58 0.76436782 0.744113030 0.5717628 285 41 815 133
159 0.025 0.59 0.77011494 0.750392465 0.5804929 278 40 822 134
160 0.025 0.60 0.77011494 0.758241758 0.7654011 268 40 832 134
161 0.025 0.61 0.77586207 0.766091052 0.8170766 259 39 841 135
162 0.025 0.62 0.78735632 0.771585557 0.6627507 254 37 846 137
163 0.025 0.63 0.79310345 0.779434851 0.7116826 245 36 855 138
164 0.025 0.64 0.80459770 0.785714286 0.5796560 239 34 861 140
165 0.025 0.65 0.81034483 0.790423862 0.5521151 234 33 866 141
166 0.025 0.66 0.81609195 0.795918367 0.5422863 228 32 872 142
167 0.025 0.67 0.81609195 0.799843014 0.6351083 223 32 877 142
168 0.025 0.68 0.82183908 0.803767661 0.5869620 219 31 881 143
169 0.025 0.69 0.82758621 0.810047096 0.5955978 212 30 888 144
170 0.025 0.70 0.83333333 0.816326531 0.6043586 205 29 895 145
171 0.025 0.71 0.83908046 0.824960754 0.6743915 195 28 905 146
172 0.025 0.72 0.83908046 0.838304553 1.0000000 178 28 922 146
173 0.025 0.73 0.83908046 0.840659341 1.0000000 175 28 925 146
174 0.025 0.74 0.85632184 0.848508634 0.8449347 168 25 932 149
175 0.025 0.75 0.85632184 0.851648352 0.9426931 164 25 936 149
176 0.025 0.76 0.86206897 0.854003140 0.8346465 162 24 938 150
177 0.025 0.77 0.86781609 0.858712716 0.7995729 157 23 943 151
178 0.025 0.78 0.87931034 0.866562009 0.6801633 149 21 951 153
179 0.025 0.79 0.87931034 0.870486656 0.8013771 144 21 956 153
180 0.025 0.80 0.88505747 0.879905808 0.9207534 133 20 967 154
181 0.025 0.81 0.89655172 0.888540031 0.8167113 124 18 976 156
182 0.025 0.82 0.89655172 0.890894819 0.8991508 121 18 979 156
183 0.025 0.83 0.90804598 0.896389325 0.6824454 116 16 984 158
184 0.025 0.84 0.91379310 0.900313972 0.6152778 112 15 988 159
185 0.025 0.85 0.91954023 0.906593407 0.6231413 105 14 995 160
186 0.025 0.86 0.92528736 0.915227630 0.7141844 95 13 1005 161
187 0.025 0.87 0.93678161 0.925431711 0.6469097 84 11 1016 163
188 0.025 0.88 0.94252874 0.932496075 0.6854239 76 10 1024 164
189 0.025 0.89 0.94252874 0.934850863 0.7822749 73 10 1027 164
190 0.025 0.90 0.94252874 0.941915228 1.0000000 64 10 1036 164
191 0.025 0.91 0.94827586 0.949764521 1.0000000 55 9 1045 165
192 0.025 0.92 0.95977011 0.955259027 0.9104711 50 7 1050 167
193 0.025 0.93 0.95977011 0.962323391 1.0000000 41 7 1059 167
194 0.025 0.94 0.97126437 0.966248038 0.8662375 38 5 1062 169
195 0.025 0.95 0.97126437 0.970172684 1.0000000 33 5 1067 169
196 0.025 0.96 0.97701149 0.975667190 1.0000000 27 4 1073 170
197 0.025 0.97 0.98275862 0.985871272 0.9770576 15 3 1085 171
198 0.025 0.98 0.98275862 0.992935636 0.2157441 6 3 1094 171
199 0.025 0.99 0.99425287 0.998430141 0.6401642 1 1 1099 173
200 0.025 1.00 1.00000000 1.000000000 0.0000000 0 0 1100 174#enrichment.plotter(h_US, "USFDR", "adj.P.Val", "FDR for Closest Upstream Hi-C Contact Overlapping Gene, Human") #These two are ugly, and can't be run anyway until next chunk is complete to create their DFs. It's OK without them.
#enrichment.plotter(c_US, "USFDR", "adj.P.Val", "FDR for Closest Upstream Hi-C Contact Overlapping Gene, Chimp")
#enrichment.plotter(gene.hic.filt, "max_B_FDR.H", "adj.P.Val", "FDR for Hi-C Contact Overlapping Gene w/ Strongest Effect Size, Human")
#enrichment.plotter(gene.hic.filt, "max_B_FDR.C", "adj.P.Val", "FDR for Hi-C Contact Overlapping Gene w/ Strongest Effect Size, Chimp")
#enrichment.plotter(gene.hic.filt, "median_FDR.H", "adj.P.Val", "Median FDR of Hi-C Contacts Overlapping Gene, Human")
#enrichment.plotter(gene.hic.filt, "median_FDR.C", "adj.P.Val", "Median FDR of Hi-C Contacts Overlapping Gene, Chimp")
enrichment.plotter(gene.hic.VC, "weighted_Z.s2post.H", "adj.P.Val", "FDR for Weighted p-val Combine of VC Hi-C Contacts Overlapping Gene", xmax=1)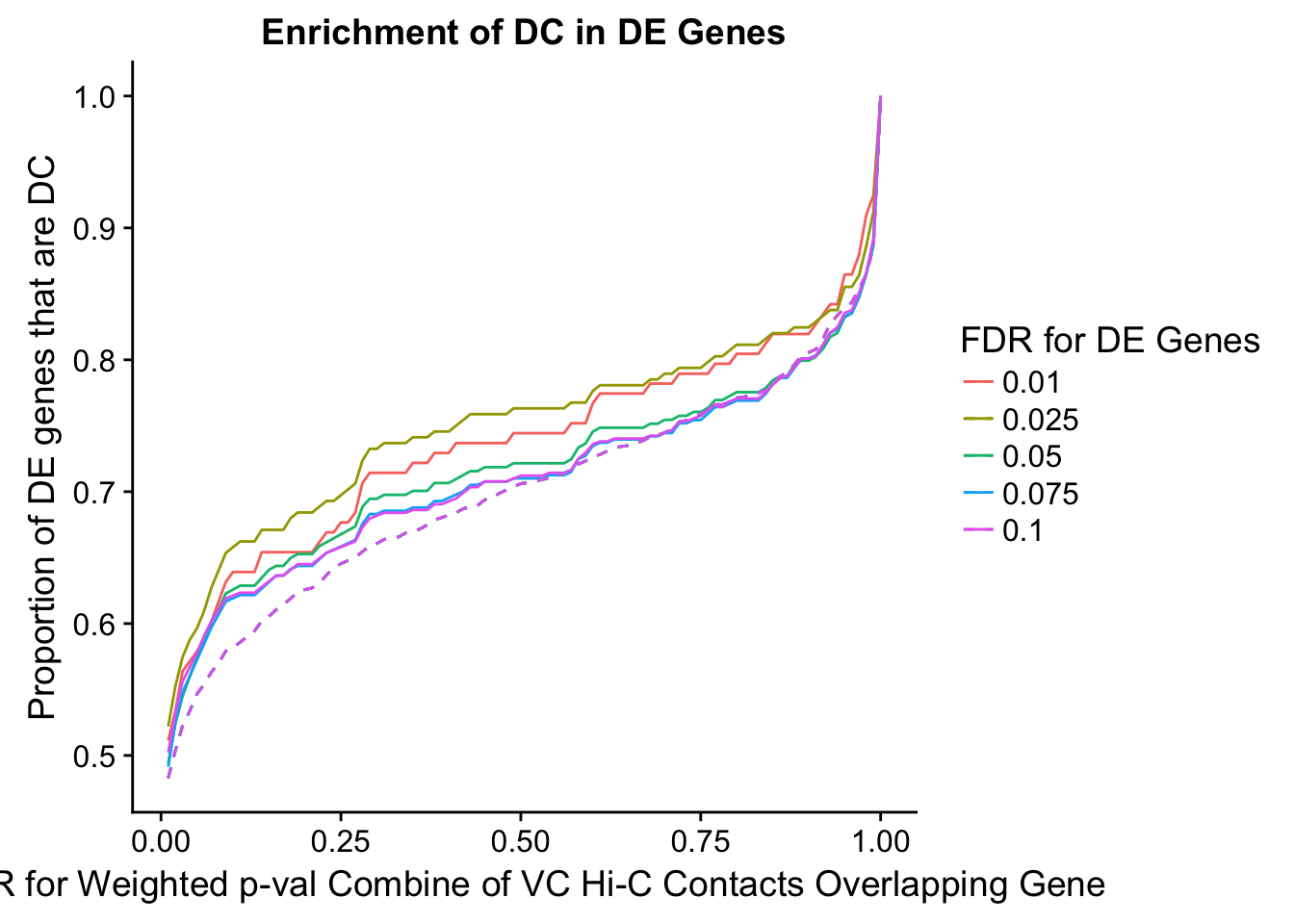
DEFDR DHICFDR prop.obs prop.exp chisq.p Dneither DE DHiC Dboth
101 0.025 0.01 0.5219298 0.4828221 0.22907489 734 109 668 119
102 0.025 0.02 0.5526316 0.5036810 0.12786275 707 102 695 126
103 0.025 0.03 0.5745614 0.5226994 0.10544398 681 97 721 131
104 0.025 0.04 0.5877193 0.5337423 0.09101518 666 94 736 134
105 0.025 0.05 0.5964912 0.5466258 0.11896633 647 92 755 136
106 0.025 0.06 0.6096491 0.5539877 0.07989392 638 89 764 139
107 0.025 0.07 0.6271930 0.5631902 0.04246358 627 85 775 143
108 0.025 0.08 0.6403509 0.5699387 0.02486832 619 82 783 146
109 0.025 0.09 0.6535088 0.5791411 0.01730370 607 79 795 149
110 0.025 0.10 0.6578947 0.5815951 0.01444978 604 78 798 150
111 0.025 0.11 0.6622807 0.5858896 0.01418556 598 77 804 151
112 0.025 0.12 0.6622807 0.5901840 0.02065730 591 77 811 151
113 0.025 0.13 0.6622807 0.5944785 0.02958599 584 77 818 151
114 0.025 0.14 0.6710526 0.6018405 0.02581156 574 75 828 153
115 0.025 0.15 0.6710526 0.6055215 0.03486092 568 75 834 153
116 0.025 0.16 0.6710526 0.6104294 0.05108028 560 75 842 153
117 0.025 0.17 0.6710526 0.6141104 0.06708667 554 75 848 153
118 0.025 0.18 0.6798246 0.6190184 0.04940579 548 73 854 155
119 0.025 0.19 0.6842105 0.6239264 0.05087720 541 72 861 156
120 0.025 0.20 0.6842105 0.6257669 0.05842344 538 72 864 156
121 0.025 0.21 0.6842105 0.6269939 0.06396079 536 72 866 156
122 0.025 0.22 0.6885965 0.6306748 0.06010761 531 71 871 157
123 0.025 0.23 0.6929825 0.6368098 0.06763219 522 70 880 158
124 0.025 0.24 0.6929825 0.6423313 0.09976023 513 70 889 158
125 0.025 0.25 0.6973684 0.6453988 0.09025385 509 69 893 159
126 0.025 0.26 0.7017544 0.6478528 0.07797052 506 68 896 160
127 0.025 0.27 0.7061404 0.6503067 0.06704398 503 67 899 161
128 0.025 0.28 0.7236842 0.6546012 0.02200172 500 63 902 165
129 0.025 0.29 0.7324561 0.6588957 0.01424774 495 61 907 167
130 0.025 0.30 0.7324561 0.6607362 0.01680808 492 61 910 167
131 0.025 0.31 0.7368421 0.6638037 0.01462045 488 60 914 168
132 0.025 0.32 0.7368421 0.6650307 0.01632663 486 60 916 168
133 0.025 0.33 0.7368421 0.6656442 0.01724451 485 60 917 168
134 0.025 0.34 0.7368421 0.6687117 0.02255725 480 60 922 168
135 0.025 0.35 0.7412281 0.6699387 0.01674004 479 59 923 169
136 0.025 0.36 0.7412281 0.6717791 0.01970266 476 59 926 169
137 0.025 0.37 0.7412281 0.6748466 0.02568183 471 59 931 169
138 0.025 0.38 0.7456140 0.6785276 0.02368458 466 58 936 170
139 0.025 0.39 0.7456140 0.6803681 0.02770891 463 58 939 170
140 0.025 0.40 0.7456140 0.6822086 0.03232147 460 58 942 170
141 0.025 0.41 0.7500000 0.6840491 0.02555596 458 57 944 171
142 0.025 0.42 0.7543860 0.6871166 0.02230723 454 56 948 172
143 0.025 0.43 0.7587719 0.6883436 0.01645828 453 55 949 173
144 0.025 0.44 0.7587719 0.6895706 0.01837320 451 55 951 173
145 0.025 0.45 0.7587719 0.6938650 0.02673086 444 55 958 173
146 0.025 0.46 0.7587719 0.6950920 0.02966572 442 55 960 173
147 0.025 0.47 0.7587719 0.6987730 0.04023061 436 55 966 173
148 0.025 0.48 0.7587719 0.7012270 0.04896639 432 55 970 173
149 0.025 0.49 0.7631579 0.7036810 0.04110648 429 54 973 174
150 0.025 0.50 0.7631579 0.7061350 0.05003213 425 54 977 174
151 0.025 0.51 0.7631579 0.7067485 0.05250793 424 54 978 174
152 0.025 0.52 0.7631579 0.7079755 0.05777576 422 54 980 174
153 0.025 0.53 0.7631579 0.7092025 0.06348788 420 54 982 174
154 0.025 0.54 0.7631579 0.7104294 0.06967225 418 54 984 174
155 0.025 0.55 0.7631579 0.7122699 0.07989711 415 54 987 174
156 0.025 0.56 0.7631579 0.7141104 0.09134851 412 54 990 174
157 0.025 0.57 0.7675439 0.7159509 0.07450337 410 53 992 175
158 0.025 0.58 0.7675439 0.7208589 0.10634338 402 53 1000 175
159 0.025 0.59 0.7675439 0.7233129 0.12603400 398 53 1004 175
160 0.025 0.60 0.7763158 0.7263804 0.08123637 395 51 1007 177
161 0.025 0.61 0.7807018 0.7282209 0.06571016 393 50 1009 178
162 0.025 0.62 0.7807018 0.7306748 0.07915871 389 50 1013 178
163 0.025 0.63 0.7807018 0.7331288 0.09484812 385 50 1017 178
164 0.025 0.64 0.7807018 0.7343558 0.10361308 383 50 1019 178
165 0.025 0.65 0.7807018 0.7349693 0.10823986 382 50 1020 178
166 0.025 0.66 0.7807018 0.7368098 0.12314653 379 50 1023 178
167 0.025 0.67 0.7807018 0.7386503 0.13967850 376 50 1026 178
168 0.025 0.68 0.7850877 0.7417178 0.12559558 372 49 1030 179
169 0.025 0.69 0.7850877 0.7429448 0.13665236 370 49 1032 179
170 0.025 0.70 0.7894737 0.7447853 0.11252583 368 48 1034 180
171 0.025 0.71 0.7894737 0.7472393 0.13359564 364 48 1038 180
172 0.025 0.72 0.7938596 0.7527607 0.14202181 356 47 1046 181
173 0.025 0.73 0.7938596 0.7539877 0.15433701 354 47 1048 181
174 0.025 0.74 0.7938596 0.7558282 0.17438696 351 47 1051 181
175 0.025 0.75 0.7938596 0.7588957 0.21227493 346 47 1056 181
176 0.025 0.76 0.7982456 0.7619632 0.19249935 342 46 1060 182
177 0.025 0.77 0.8026316 0.7644172 0.16696881 339 45 1063 183
178 0.025 0.78 0.8026316 0.7644172 0.16696881 339 45 1063 183
179 0.025 0.79 0.8070175 0.7674847 0.15011400 335 44 1067 184
180 0.025 0.80 0.8114035 0.7711656 0.14034315 330 43 1072 185
181 0.025 0.81 0.8114035 0.7717791 0.14647156 329 43 1073 185
182 0.025 0.82 0.8114035 0.7742331 0.17316659 325 43 1077 185
183 0.025 0.83 0.8114035 0.7748466 0.18040810 324 43 1078 185
184 0.025 0.84 0.8157895 0.7766871 0.14904141 322 42 1080 186
185 0.025 0.85 0.8201754 0.7809816 0.14522864 316 41 1086 187
186 0.025 0.86 0.8201754 0.7871166 0.21957730 306 41 1096 187
187 0.025 0.87 0.8201754 0.7907975 0.27651749 300 41 1102 187
188 0.025 0.88 0.8245614 0.7963190 0.29229895 292 40 1110 188
189 0.025 0.89 0.8245614 0.8030675 0.42941752 281 40 1121 188
190 0.025 0.90 0.8245614 0.8055215 0.48830870 277 40 1125 188
191 0.025 0.91 0.8289474 0.8079755 0.43762365 274 39 1128 189
192 0.025 0.92 0.8333333 0.8159509 0.52336925 262 38 1140 190
193 0.025 0.93 0.8377193 0.8276074 0.73284982 244 37 1158 191
194 0.025 0.94 0.8377193 0.8337423 0.93781706 234 37 1168 191
195 0.025 0.95 0.8552632 0.8392638 0.54052911 229 33 1173 195
196 0.025 0.96 0.8552632 0.8435583 0.66988567 222 33 1180 195
197 0.025 0.97 0.8640351 0.8539877 0.71724625 207 31 1195 197
198 0.025 0.98 0.8859649 0.8644172 0.35732592 195 26 1207 202
199 0.025 0.99 0.9122807 0.8858896 0.21529733 166 20 1236 208
200 0.025 1.00 1.0000000 1.0000000 0.00000000 0 0 1402 228enrichment.plotter(gene.hic.KR, "weighted_Z.s2post.H", "adj.P.Val", "FDR for Weighted p-val Combine of KR Hi-C Contacts Overlapping Gene", xmax=1)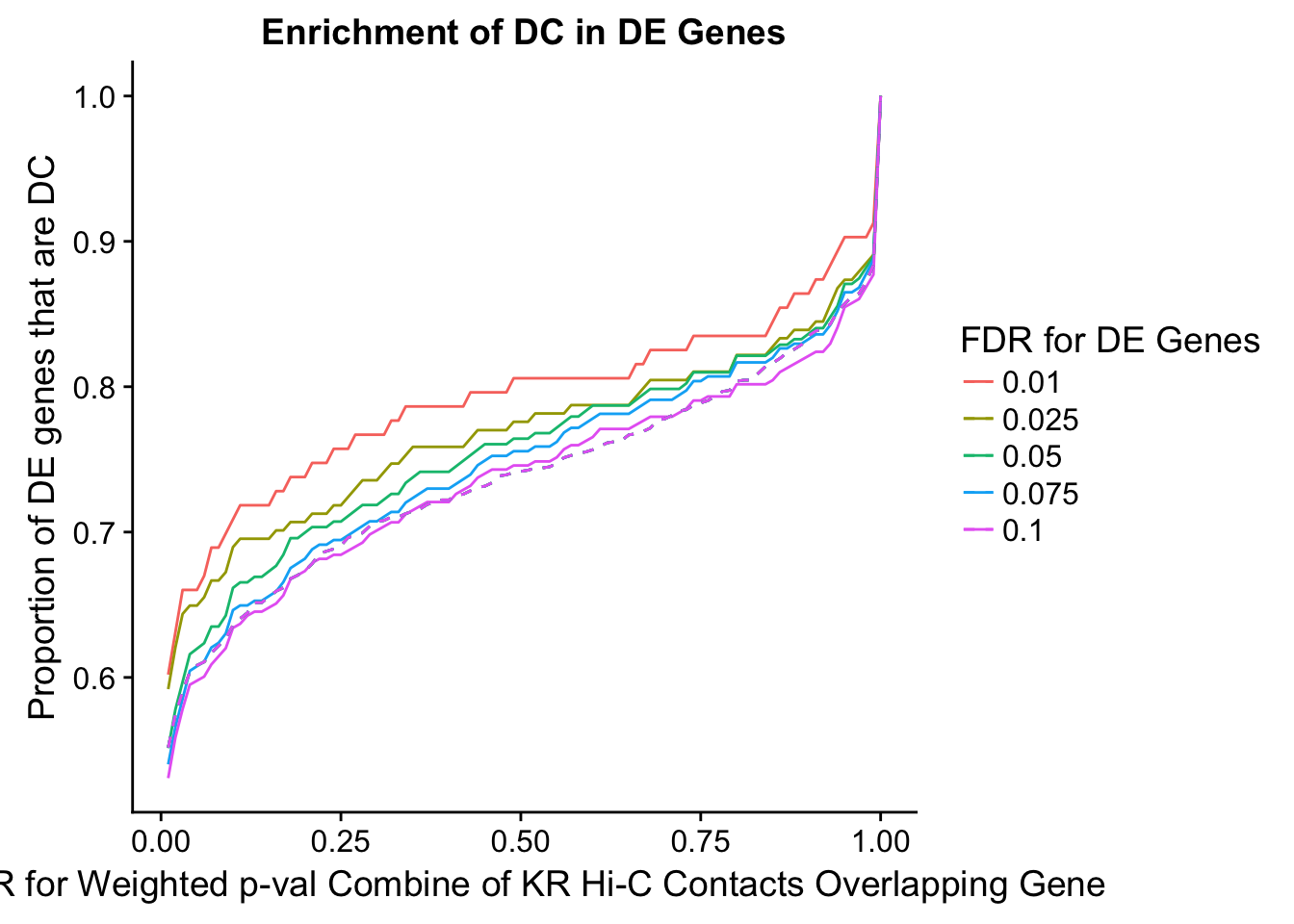
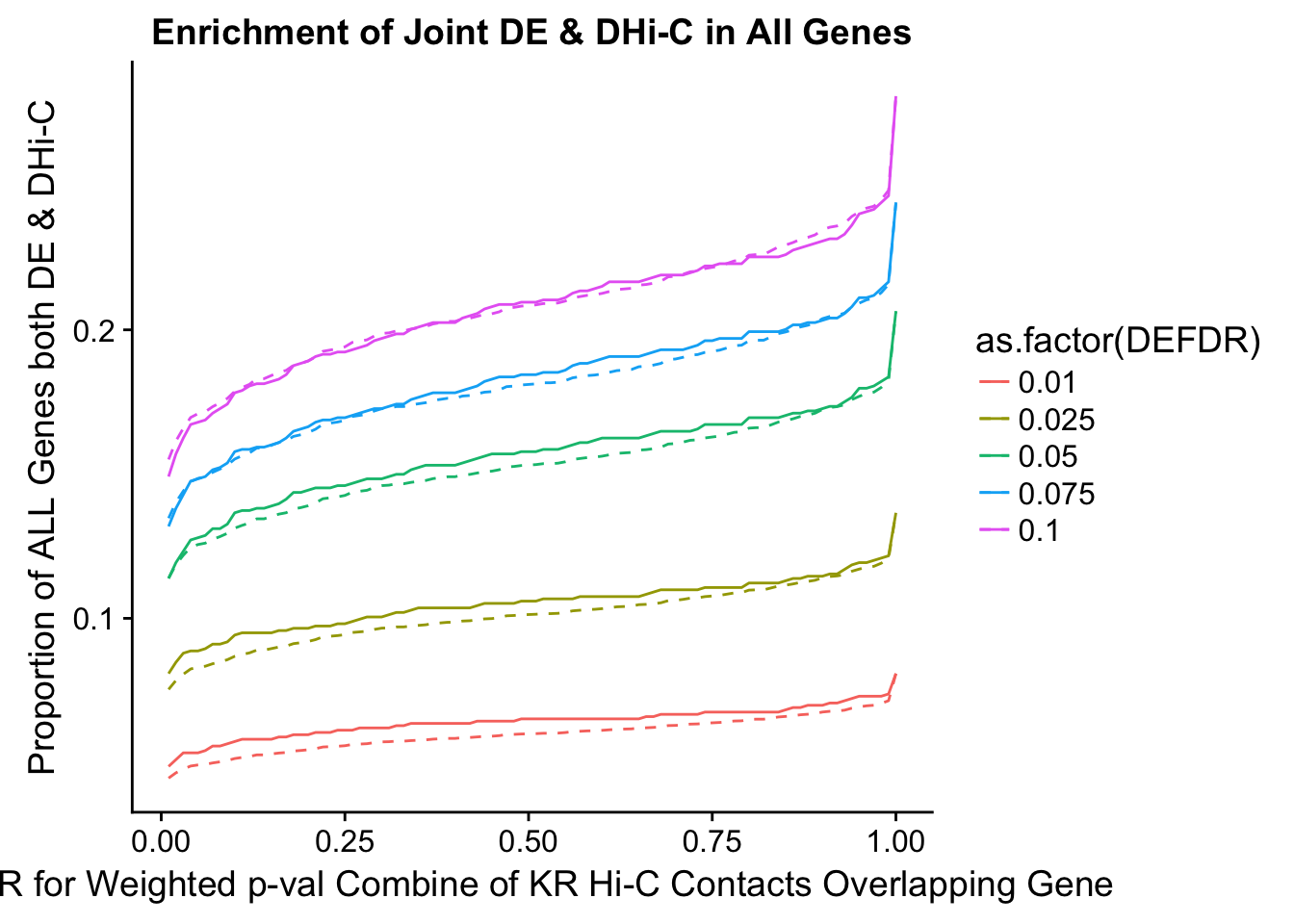
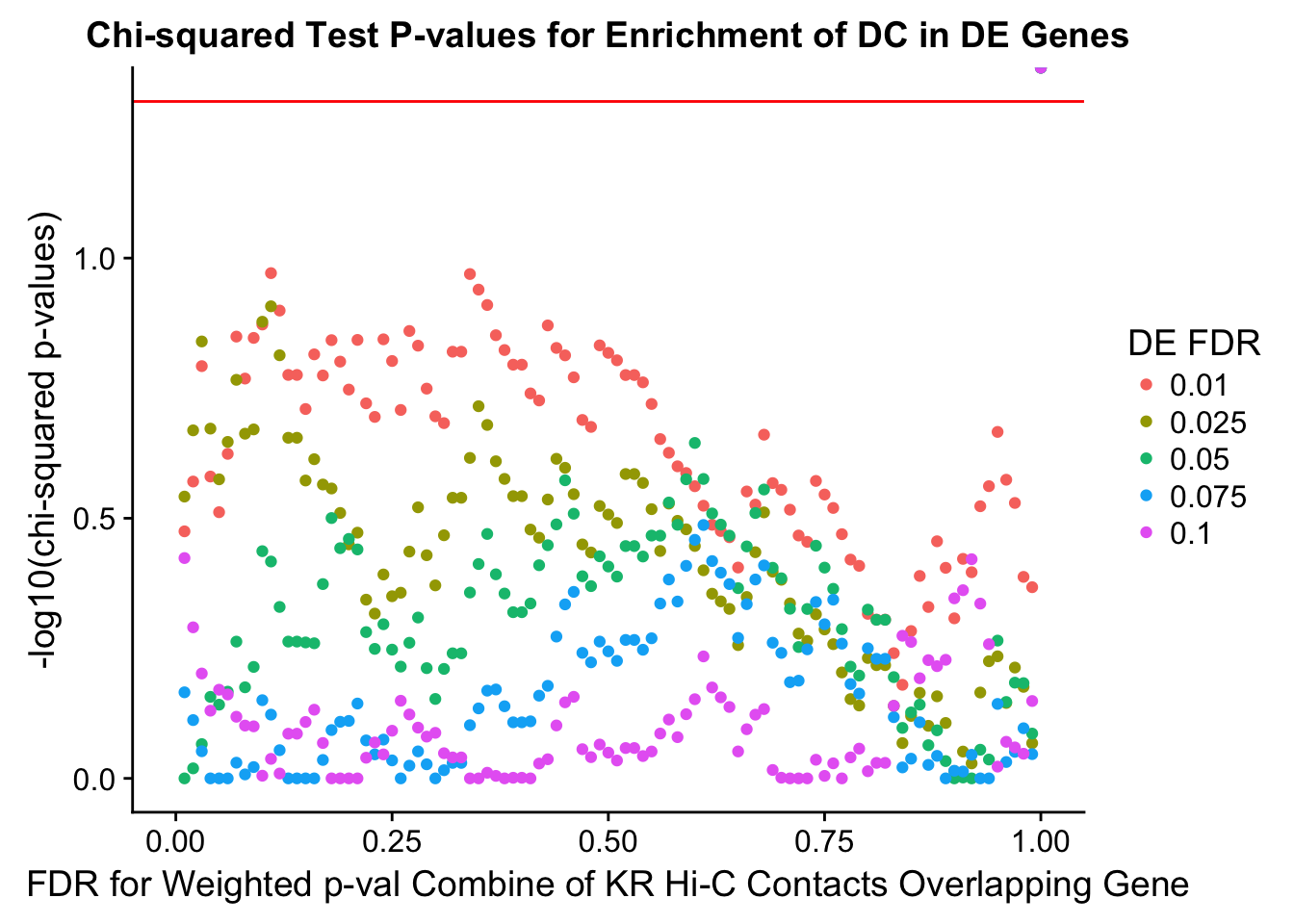
DEFDR DHICFDR prop.obs prop.exp chisq.p Dneither DE DHiC Dboth
101 0.025 0.01 0.5919540 0.5518053 0.2873130 500 71 600 103
102 0.025 0.02 0.6206897 0.5745683 0.2143213 476 66 624 108
103 0.025 0.03 0.6436782 0.5902669 0.1446126 460 62 640 112
104 0.025 0.04 0.6494253 0.6036107 0.2126821 444 61 656 113
105 0.025 0.05 0.6494253 0.6083203 0.2661959 438 61 662 113
106 0.025 0.06 0.6551724 0.6106750 0.2255754 436 60 664 114
107 0.025 0.07 0.6666667 0.6169545 0.1713818 430 58 670 116
108 0.025 0.08 0.6666667 0.6216641 0.2175070 424 58 676 116
109 0.025 0.09 0.6724138 0.6271586 0.2134258 418 57 682 117
110 0.025 0.10 0.6896552 0.6357928 0.1325295 410 54 690 120
111 0.025 0.11 0.6954023 0.6405024 0.1237700 405 53 695 121
112 0.025 0.12 0.6954023 0.6444270 0.1537234 400 53 700 121
113 0.025 0.13 0.6954023 0.6514914 0.2214840 391 53 709 121
114 0.025 0.14 0.6954023 0.6514914 0.2214840 391 53 709 121
115 0.025 0.15 0.6954023 0.6554160 0.2675974 386 53 714 121
116 0.025 0.16 0.7011494 0.6593407 0.2435148 382 52 718 122
117 0.025 0.17 0.7011494 0.6616954 0.2723963 379 52 721 122
118 0.025 0.18 0.7068966 0.6679749 0.2772023 372 51 728 123
119 0.025 0.19 0.7068966 0.6703297 0.3089288 369 51 731 123
120 0.025 0.20 0.7068966 0.6734694 0.3550071 365 51 735 123
121 0.025 0.21 0.7126437 0.6781790 0.3370799 360 50 740 124
122 0.025 0.22 0.7126437 0.6852433 0.4534289 351 50 749 124
123 0.025 0.23 0.7126437 0.6868132 0.4822571 349 50 751 124
124 0.025 0.24 0.7183908 0.6883830 0.4055917 348 49 752 125
125 0.025 0.25 0.7183908 0.6907378 0.4466055 345 49 755 125
126 0.025 0.26 0.7241379 0.6962323 0.4396997 339 48 761 126
127 0.025 0.27 0.7298851 0.6978022 0.3665430 338 47 762 127
128 0.025 0.28 0.7356322 0.6993721 0.3013083 337 46 763 128
129 0.025 0.29 0.7356322 0.7040816 0.3724654 331 46 769 128
130 0.025 0.30 0.7356322 0.7072214 0.4256303 327 46 773 128
131 0.025 0.31 0.7413793 0.7080063 0.3409714 327 45 773 129
132 0.025 0.32 0.7471264 0.7103611 0.2888295 325 44 775 130
133 0.025 0.33 0.7471264 0.7103611 0.2888295 325 44 775 130
134 0.025 0.34 0.7528736 0.7127159 0.2421232 323 43 777 131
135 0.025 0.35 0.7586207 0.7142857 0.1926150 322 42 778 132
136 0.025 0.36 0.7586207 0.7158556 0.2092502 320 42 780 132
137 0.025 0.37 0.7586207 0.7189953 0.2457615 316 42 784 132
138 0.025 0.38 0.7586207 0.7205651 0.2656961 314 42 786 132
139 0.025 0.39 0.7586207 0.7221350 0.2867836 312 42 788 132
140 0.025 0.40 0.7586207 0.7221350 0.2867836 312 42 788 132
141 0.025 0.41 0.7586207 0.7252747 0.3324947 308 42 792 132
142 0.025 0.42 0.7586207 0.7260597 0.3446697 307 42 793 132
143 0.025 0.43 0.7643678 0.7284144 0.2910577 305 41 795 133
144 0.025 0.44 0.7701149 0.7307692 0.2431008 303 40 797 134
145 0.025 0.45 0.7701149 0.7315542 0.2529535 302 40 798 134
146 0.025 0.46 0.7701149 0.7339089 0.2842756 299 40 801 134
147 0.025 0.47 0.7701149 0.7386185 0.3550914 293 40 807 134
148 0.025 0.48 0.7701149 0.7394035 0.3679744 292 40 808 134
149 0.025 0.49 0.7758621 0.7409733 0.2995502 291 39 809 135
150 0.025 0.50 0.7758621 0.7417582 0.3110771 290 39 810 135
151 0.025 0.51 0.7758621 0.7425432 0.3229145 289 39 811 135
152 0.025 0.52 0.7816092 0.7441130 0.2600110 288 38 812 136
153 0.025 0.53 0.7816092 0.7441130 0.2600110 288 38 812 136
154 0.025 0.54 0.7816092 0.7448980 0.2704876 287 38 813 136
155 0.025 0.55 0.7816092 0.7472527 0.3037634 284 38 816 136
156 0.025 0.56 0.7816092 0.7511774 0.3655209 279 38 821 136
157 0.025 0.57 0.7873563 0.7527473 0.2963600 278 37 822 137
158 0.025 0.58 0.7873563 0.7543171 0.3198609 276 37 824 137
159 0.025 0.59 0.7873563 0.7551020 0.3320930 275 37 825 137
160 0.025 0.60 0.7873563 0.7566719 0.3575289 273 37 827 137
161 0.025 0.61 0.7873563 0.7590267 0.3981275 270 37 830 137
162 0.025 0.62 0.7873563 0.7613815 0.4416625 267 37 833 137
163 0.025 0.63 0.7873563 0.7621664 0.4568228 266 37 834 137
164 0.025 0.64 0.7873563 0.7629513 0.4723044 265 37 835 137
165 0.025 0.65 0.7873563 0.7668760 0.5544299 260 37 840 137
166 0.025 0.66 0.7931034 0.7676609 0.4480750 260 36 840 138
167 0.025 0.67 0.7988506 0.7692308 0.3674960 259 35 841 139
168 0.025 0.68 0.8045977 0.7715856 0.3081386 257 34 843 140
169 0.025 0.69 0.8045977 0.7770801 0.4005951 250 34 850 140
170 0.025 0.70 0.8045977 0.7778650 0.4151801 249 34 851 140
171 0.025 0.71 0.8045977 0.7802198 0.4609993 246 34 854 140
172 0.025 0.72 0.8045977 0.7833595 0.5268371 242 34 858 140
173 0.025 0.73 0.8045977 0.7841444 0.5441232 241 34 859 140
174 0.025 0.74 0.8103448 0.7872841 0.4837525 238 33 862 141
175 0.025 0.75 0.8103448 0.7888540 0.5172609 236 33 864 141
176 0.025 0.76 0.8103448 0.7904239 0.5521151 234 33 866 141
177 0.025 0.77 0.8103448 0.7935636 0.6256979 230 33 870 141
178 0.025 0.78 0.8103448 0.7967033 0.7040755 226 33 874 141
179 0.025 0.79 0.8103448 0.7974882 0.7243542 225 33 875 141
180 0.025 0.80 0.8218391 0.8037677 0.5869620 219 31 881 143
181 0.025 0.81 0.8218391 0.8045526 0.6058766 218 31 882 143
182 0.025 0.82 0.8218391 0.8045526 0.6058766 218 31 882 143
183 0.025 0.83 0.8218391 0.8092622 0.7258825 212 31 888 143
184 0.025 0.84 0.8218391 0.8139717 0.8554424 206 31 894 143
185 0.025 0.85 0.8275862 0.8163265 0.7585041 204 30 896 144
186 0.025 0.86 0.8333333 0.8194662 0.6849286 201 29 899 145
187 0.025 0.87 0.8333333 0.8233909 0.7924348 196 29 904 145
188 0.025 0.88 0.8390805 0.8257457 0.6954288 194 28 906 146
189 0.025 0.89 0.8390805 0.8288854 0.7825661 190 28 910 146
190 0.025 0.90 0.8390805 0.8351648 0.9681968 182 28 918 146
191 0.025 0.91 0.8448276 0.8383046 0.8880944 179 27 921 147
192 0.025 0.92 0.8448276 0.8398744 0.9358379 177 27 923 147
193 0.025 0.93 0.8563218 0.8430141 0.6838856 175 25 925 149
194 0.025 0.94 0.8678161 0.8516484 0.5954581 166 23 934 151
195 0.025 0.95 0.8735632 0.8571429 0.5826152 160 22 940 152
196 0.025 0.96 0.8735632 0.8618524 0.7161775 154 22 946 152
197 0.025 0.97 0.8793103 0.8642072 0.6123034 152 21 948 153
198 0.025 0.98 0.8850575 0.8720565 0.6668997 143 20 957 154
199 0.025 0.99 0.8908046 0.8838305 0.8558436 129 19 971 155
200 0.025 1.00 1.0000000 1.0000000 0.0000000 0 0 1100 174enrichment.plotter(gene.hic.VC, "weighted_Z.ALLvar.H", "adj.P.Val", "FDR for Weighted p-val Combine of VC Hi-C Contacts Overlapping Gene", xmax=1)Warning in chisq.test(mytable): Chi-squared approximation may be incorrectWarning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect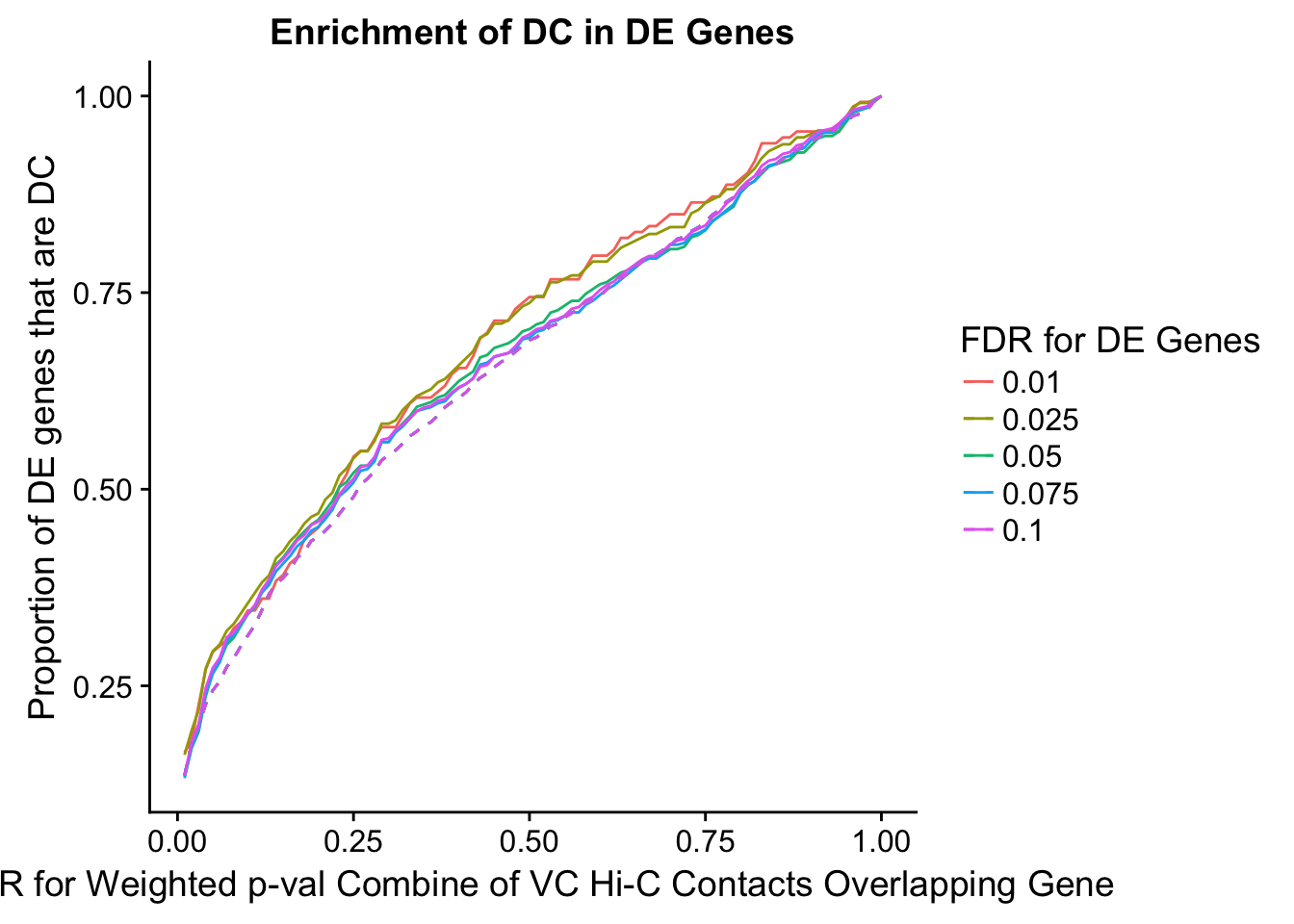
DEFDR DHICFDR prop.obs prop.exp chisq.p Dneither DE DHiC Dboth
101 0.025 0.01 0.1622807 0.1361963 0.25676655 1217 191 185 37
102 0.025 0.02 0.1929825 0.1766871 0.54717641 1158 184 244 44
103 0.025 0.03 0.2192982 0.1993865 0.47026993 1127 178 275 50
104 0.025 0.04 0.2719298 0.2263804 0.09164595 1095 166 307 62
105 0.025 0.05 0.2938596 0.2441718 0.07185914 1071 161 331 67
106 0.025 0.06 0.3026316 0.2558282 0.09599076 1054 159 348 69
107 0.025 0.07 0.3201754 0.2742331 0.11035166 1028 155 374 73
108 0.025 0.08 0.3289474 0.2852761 0.13476600 1012 153 390 75
109 0.025 0.09 0.3421053 0.3006135 0.16289065 990 150 412 78
110 0.025 0.10 0.3552632 0.3141104 0.17175699 971 147 431 81
111 0.025 0.11 0.3684211 0.3276074 0.18033127 952 144 450 84
112 0.025 0.12 0.3815789 0.3466258 0.26237939 924 141 478 87
113 0.025 0.13 0.3903509 0.3668712 0.47207520 893 139 509 89
114 0.025 0.14 0.4122807 0.3791411 0.29904069 878 134 524 94
115 0.025 0.15 0.4210526 0.3871166 0.28867753 867 132 535 96
116 0.025 0.16 0.4342105 0.3975460 0.25145941 853 129 549 99
117 0.025 0.17 0.4429825 0.4128834 0.35610932 830 127 572 101
118 0.025 0.18 0.4561404 0.4202454 0.26628774 821 124 581 104
119 0.025 0.19 0.4649123 0.4343558 0.35151820 800 122 602 106
120 0.025 0.20 0.4692982 0.4392638 0.36105852 793 121 609 107
121 0.025 0.21 0.4868421 0.4472393 0.22057666 784 117 618 111
122 0.025 0.22 0.4956140 0.4564417 0.22676521 771 115 631 113
123 0.025 0.23 0.5175439 0.4687117 0.12809178 756 110 646 118
124 0.025 0.24 0.5263158 0.4803681 0.15390862 739 108 663 120
125 0.025 0.25 0.5394737 0.4901840 0.12505954 726 105 676 123
126 0.025 0.26 0.5482456 0.5061350 0.19363005 702 103 700 125
127 0.025 0.27 0.5482456 0.5134969 0.28892624 690 103 712 125
128 0.025 0.28 0.5614035 0.5233129 0.24192522 677 100 725 128
129 0.025 0.29 0.5833333 0.5368098 0.14777399 660 95 742 133
130 0.025 0.30 0.5833333 0.5435583 0.21928372 649 95 753 133
131 0.025 0.31 0.5877193 0.5496933 0.24094883 640 94 762 134
132 0.025 0.32 0.6008772 0.5582822 0.18529777 629 91 773 137
133 0.025 0.33 0.6096491 0.5674847 0.18898445 616 89 786 139
134 0.025 0.34 0.6184211 0.5736196 0.16069915 608 87 794 141
135 0.025 0.35 0.6228070 0.5809816 0.19094222 597 86 805 142
136 0.025 0.36 0.6271930 0.5852761 0.18926982 591 85 811 143
137 0.025 0.37 0.6359649 0.5938650 0.18583914 579 83 823 145
138 0.025 0.38 0.6403509 0.6042945 0.25954094 563 82 839 146
139 0.025 0.39 0.6491228 0.6110429 0.23072437 554 80 848 148
140 0.025 0.40 0.6578947 0.6165644 0.19002059 547 78 855 150
141 0.025 0.41 0.6666667 0.6233129 0.16665957 538 76 864 152
142 0.025 0.42 0.6754386 0.6331288 0.17534683 524 74 878 154
143 0.025 0.43 0.6929825 0.6417178 0.09566872 514 70 888 158
144 0.025 0.44 0.6973684 0.6472393 0.10239636 506 69 896 159
145 0.025 0.45 0.7105263 0.6552147 0.06882517 496 66 906 162
146 0.025 0.46 0.7105263 0.6619632 0.11049384 485 66 917 162
147 0.025 0.47 0.7149123 0.6668712 0.11325464 478 65 924 163
148 0.025 0.48 0.7236842 0.6748466 0.10497047 467 63 935 165
149 0.025 0.49 0.7324561 0.6822086 0.09289489 457 61 945 167
150 0.025 0.50 0.7368421 0.6895706 0.11267026 446 60 956 168
151 0.025 0.51 0.7456140 0.6932515 0.07651190 442 58 960 170
152 0.025 0.52 0.7456140 0.6993865 0.11791809 432 58 970 170
153 0.025 0.53 0.7631579 0.7073620 0.05508802 423 54 979 174
154 0.025 0.54 0.7631579 0.7104294 0.06967225 418 54 984 174
155 0.025 0.55 0.7675439 0.7184049 0.08924920 406 53 996 175
156 0.025 0.56 0.7719298 0.7239264 0.09524241 398 52 1004 176
157 0.025 0.57 0.7719298 0.7300613 0.14563742 388 52 1014 176
158 0.025 0.58 0.7807018 0.7368098 0.12314653 379 50 1023 178
159 0.025 0.59 0.7894737 0.7411043 0.08609842 374 48 1028 180
160 0.025 0.60 0.7894737 0.7453988 0.11751957 367 48 1035 180
161 0.025 0.61 0.7894737 0.7546012 0.21630109 352 48 1050 180
162 0.025 0.62 0.7982456 0.7619632 0.19249935 342 46 1060 182
163 0.025 0.63 0.8070175 0.7687117 0.16319330 333 44 1069 184
164 0.025 0.64 0.8114035 0.7742331 0.17316659 325 43 1077 185
165 0.025 0.65 0.8157895 0.7822086 0.21566617 313 42 1089 186
166 0.025 0.66 0.8201754 0.7895706 0.25643777 302 41 1100 187
167 0.025 0.67 0.8245614 0.7957055 0.28161943 293 40 1109 188
168 0.025 0.68 0.8245614 0.7987730 0.33795762 288 40 1114 188
169 0.025 0.69 0.8289474 0.8042945 0.35668894 280 39 1122 189
170 0.025 0.70 0.8333333 0.8098160 0.37633186 272 38 1130 190
171 0.025 0.71 0.8333333 0.8184049 0.59067725 258 38 1144 190
172 0.025 0.72 0.8333333 0.8208589 0.66245528 254 38 1148 190
173 0.025 0.73 0.8508772 0.8282209 0.37707699 246 34 1156 194
174 0.025 0.74 0.8552632 0.8331288 0.38388918 239 33 1163 195
175 0.025 0.75 0.8640351 0.8404908 0.34241374 229 31 1173 197
176 0.025 0.76 0.8684211 0.8490798 0.43542672 216 30 1186 198
177 0.025 0.77 0.8728070 0.8552147 0.47615129 207 29 1195 199
178 0.025 0.78 0.8815789 0.8656442 0.51179611 192 27 1210 201
179 0.025 0.79 0.8815789 0.8711656 0.68952958 183 27 1219 201
180 0.025 0.80 0.8903509 0.8809816 0.71822985 169 25 1233 203
181 0.025 0.81 0.8991228 0.8877301 0.63517810 160 23 1242 205
182 0.025 0.82 0.9078947 0.8944785 0.55199675 151 21 1251 207
183 0.025 0.83 0.9210526 0.9042945 0.42019543 138 18 1264 210
184 0.025 0.84 0.9298246 0.9092025 0.29634739 132 16 1270 212
185 0.025 0.85 0.9342105 0.9128834 0.26929726 127 15 1275 213
186 0.025 0.86 0.9385965 0.9202454 0.33151616 116 14 1286 214
187 0.025 0.87 0.9385965 0.9251534 0.48638596 108 14 1294 214
188 0.025 0.88 0.9473684 0.9319018 0.39096247 99 12 1303 216
189 0.025 0.89 0.9473684 0.9374233 0.60228642 90 12 1312 216
190 0.025 0.90 0.9517544 0.9423313 0.61358405 83 11 1319 217
191 0.025 0.91 0.9561404 0.9472393 0.62516609 76 10 1326 218
192 0.025 0.92 0.9561404 0.9533742 0.96469811 66 10 1336 218
193 0.025 0.93 0.9561404 0.9546012 1.00000000 64 10 1338 218
194 0.025 0.94 0.9649123 0.9625767 0.99023930 53 8 1349 220
195 0.025 0.95 0.9736842 0.9674847 0.71303401 47 6 1355 222
196 0.025 0.96 0.9868421 0.9748466 0.30810997 38 3 1364 225
197 0.025 0.97 0.9912281 0.9773006 0.19959242 35 2 1367 226
198 0.025 0.98 0.9912281 0.9822086 0.40047575 27 2 1375 226
199 0.025 0.99 0.9956140 0.9938650 1.00000000 9 1 1393 227
200 0.025 1.00 1.0000000 1.0000000 0.00000000 0 0 1402 228enrichment.plotter(gene.hic.KR, "weighted_Z.ALLvar.H", "adj.P.Val", "FDR for Weighted p-val Combine of KR Hi-C Contacts Overlapping Gene", xmax=1)Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect
Warning in chisq.test(mytable): Chi-squared approximation may be incorrect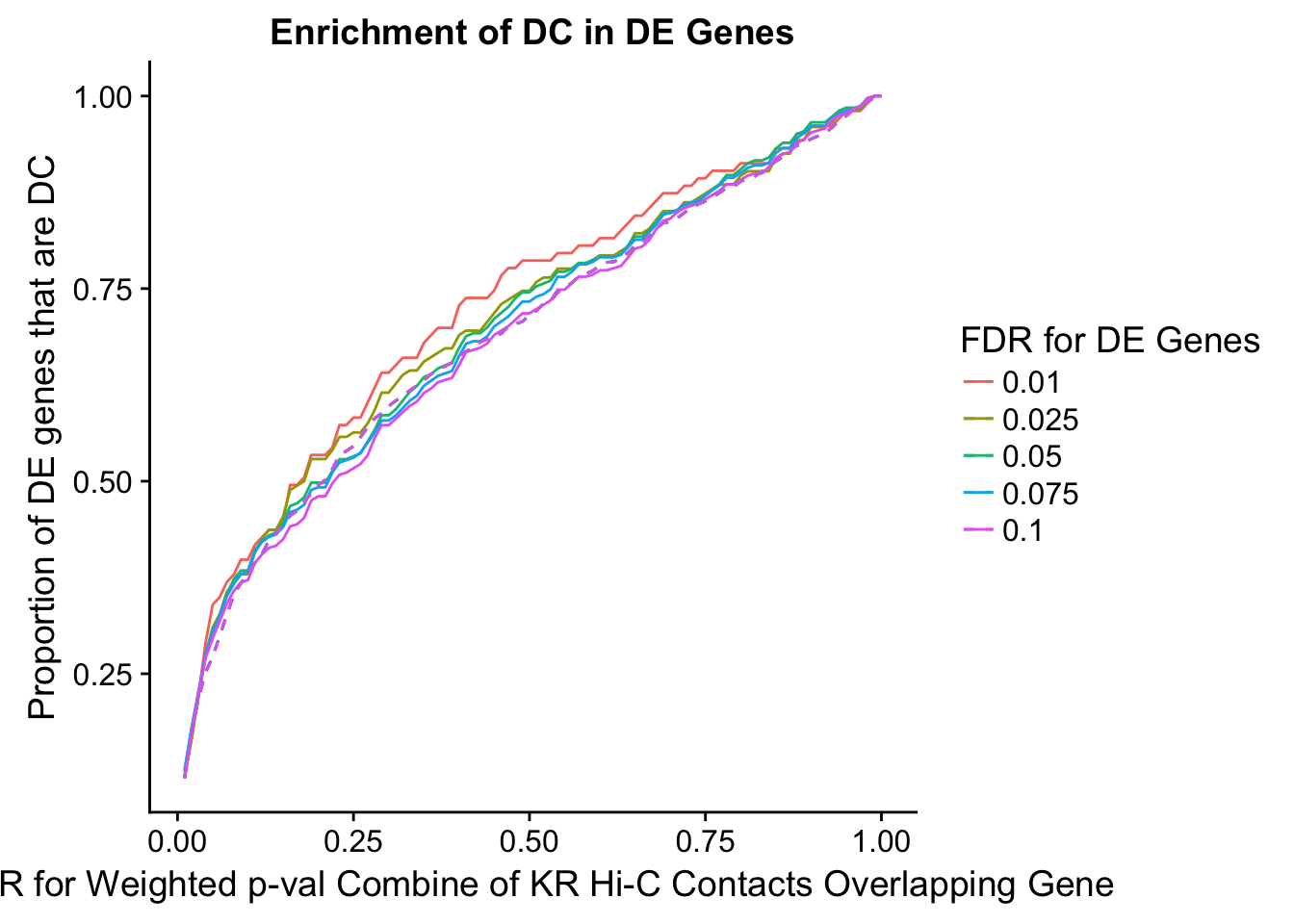
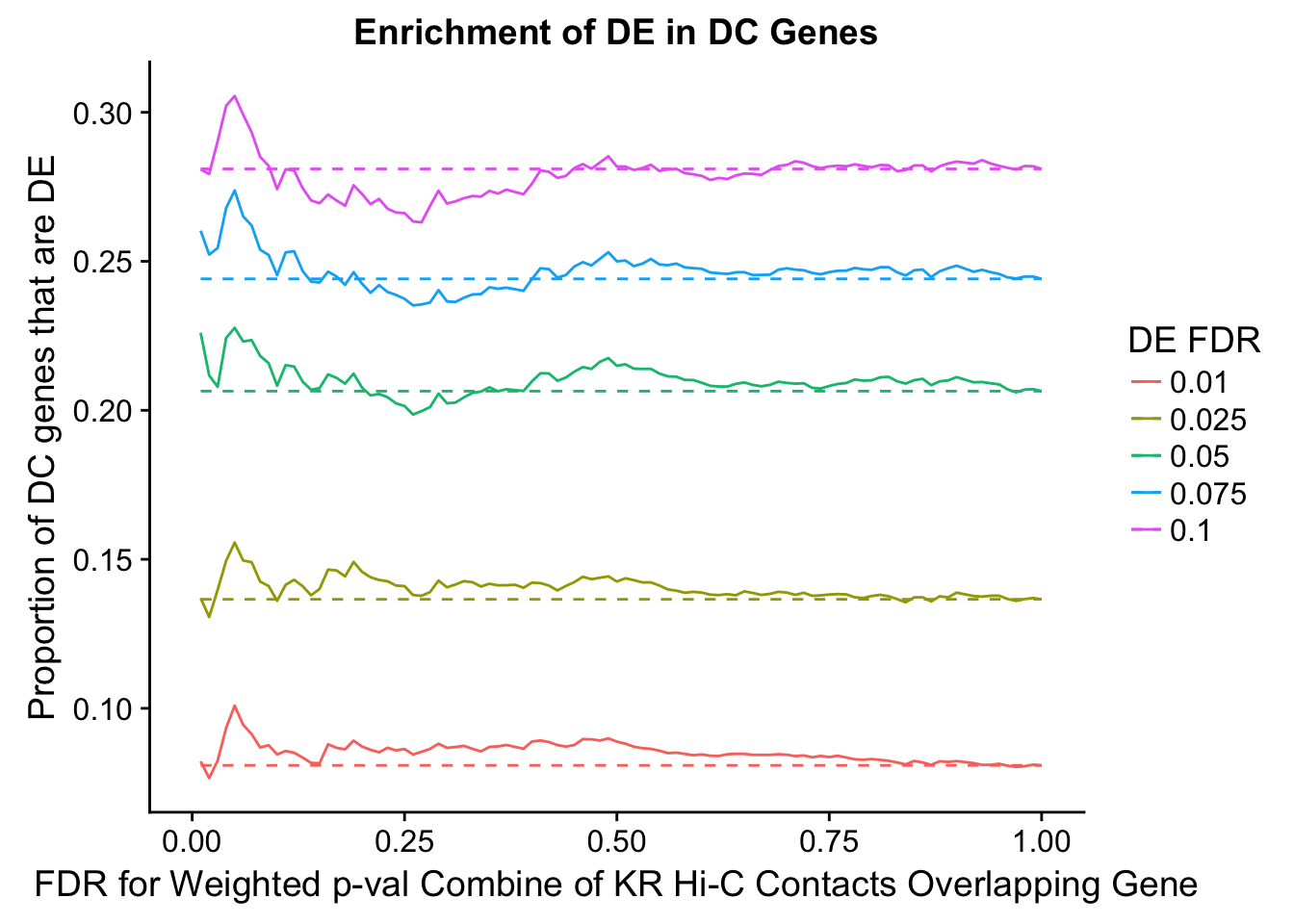
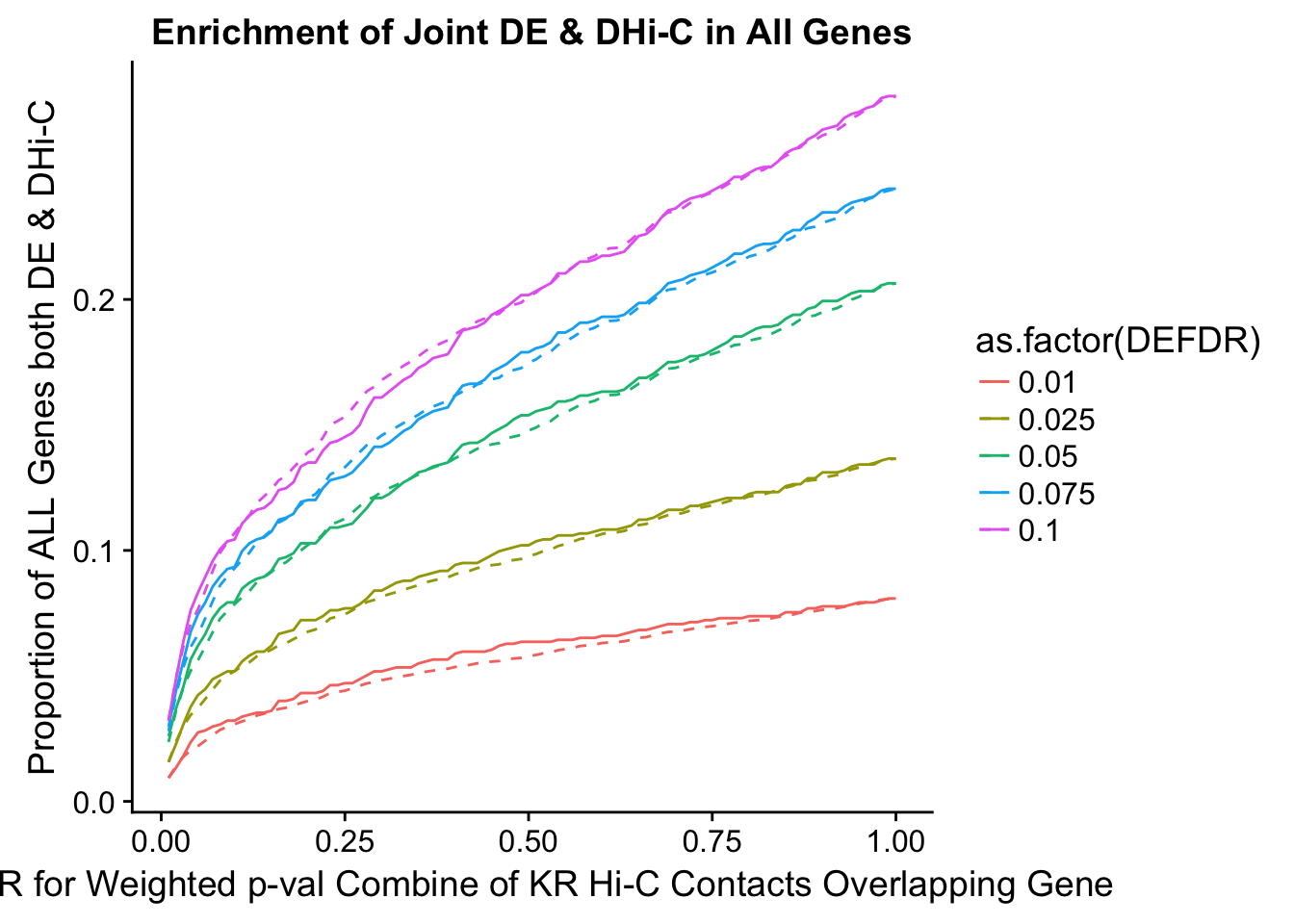
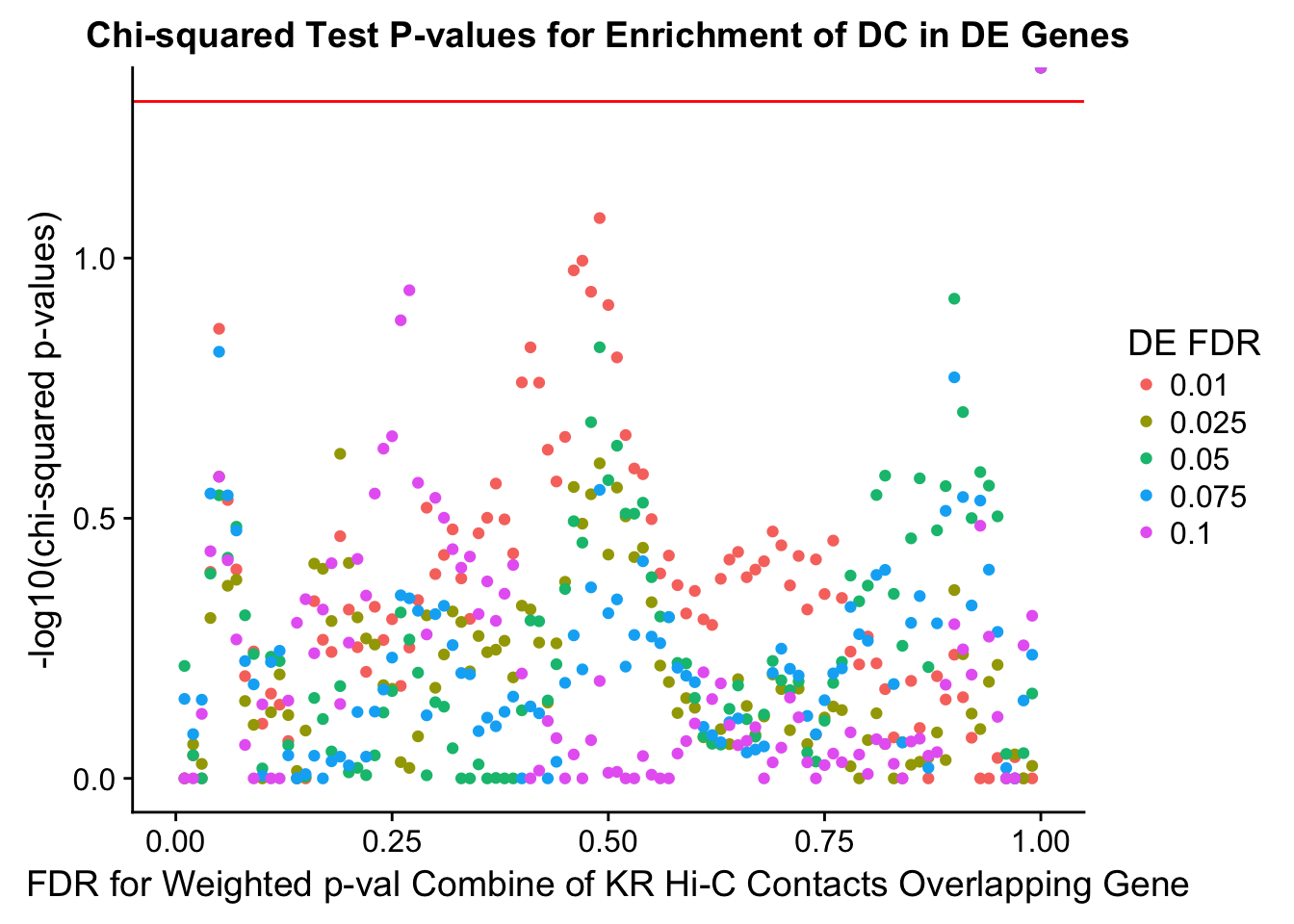
DEFDR DHICFDR prop.obs prop.exp chisq.p Dneither DE DHiC Dboth
101 0.025 0.01 0.1149425 0.1145997 1.0000000 974 154 126 20
102 0.025 0.02 0.1666667 0.1742543 0.8599647 907 145 193 29
103 0.025 0.03 0.2241379 0.2189953 0.9379175 860 135 240 39
104 0.025 0.04 0.2758621 0.2519623 0.4917466 827 126 273 48
105 0.025 0.05 0.3103448 0.2723705 0.2630143 807 120 293 54
106 0.025 0.06 0.3275862 0.2990581 0.4263562 776 117 324 57
107 0.025 0.07 0.3563218 0.3265306 0.4151562 746 112 354 62
108 0.025 0.08 0.3678161 0.3524333 0.7101029 715 110 385 64
109 0.025 0.09 0.3793103 0.3673469 0.7889540 698 108 402 66
110 0.025 0.10 0.3793103 0.3806907 1.0000000 681 108 419 66
111 0.025 0.11 0.4080460 0.3940345 0.7462602 669 103 431 71
112 0.025 0.12 0.4252874 0.4058085 0.6311919 657 100 443 74
113 0.025 0.13 0.4367816 0.4230769 0.7556332 637 98 463 76
114 0.025 0.14 0.4367816 0.4324961 0.9677273 625 98 475 76
115 0.025 0.15 0.4540230 0.4427002 0.8091824 615 95 485 79
116 0.025 0.16 0.4885057 0.4552590 0.3865878 605 89 495 85
117 0.025 0.17 0.4942529 0.4615385 0.3954622 598 88 502 86
118 0.025 0.18 0.5000000 0.4733124 0.4983497 584 87 516 87
119 0.025 0.19 0.5287356 0.4843014 0.2377760 575 82 525 92
120 0.025 0.20 0.5287356 0.4952904 0.3853819 561 82 539 92
121 0.025 0.21 0.5287356 0.5015699 0.4903809 553 82 547 92
122 0.025 0.22 0.5402299 0.5156986 0.5384190 537 80 563 94
123 0.025 0.23 0.5574713 0.5337520 0.5530465 517 77 583 97
124 0.025 0.24 0.5574713 0.5392465 0.6619677 510 77 590 97
125 0.025 0.25 0.5632184 0.5455259 0.6726667 503 76 597 98
126 0.025 0.26 0.5632184 0.5572998 0.9306509 488 76 612 98
127 0.025 0.27 0.5747126 0.5698587 0.9547179 474 74 626 100
128 0.025 0.28 0.5919540 0.5816327 0.8302877 462 71 638 103
129 0.025 0.29 0.6149425 0.5879121 0.4859837 458 67 642 107
130 0.025 0.30 0.6149425 0.5973312 0.6696771 446 67 654 107
131 0.025 0.31 0.6264368 0.6043956 0.5778913 439 65 661 109
132 0.025 0.32 0.6379310 0.6106750 0.4777848 433 63 667 111
133 0.025 0.33 0.6436782 0.6177394 0.5004319 425 62 675 112
134 0.025 0.34 0.6436782 0.6240188 0.6227538 417 62 683 112
135 0.025 0.35 0.6551724 0.6310832 0.5325076 410 60 690 114
136 0.025 0.36 0.6609195 0.6389325 0.5721334 401 59 699 115
137 0.025 0.37 0.6666667 0.6444270 0.5657513 395 58 705 116
138 0.025 0.38 0.6724138 0.6491366 0.5439018 390 57 710 117
139 0.025 0.39 0.6724138 0.6538462 0.6395684 384 57 716 117
140 0.025 0.40 0.6896552 0.6624804 0.4656651 376 54 724 120
141 0.025 0.41 0.6954023 0.6687598 0.4734283 369 53 731 121
142 0.025 0.42 0.6954023 0.6726845 0.5482686 364 53 736 121
143 0.025 0.43 0.6954023 0.6805338 0.7149655 354 53 746 121
144 0.025 0.44 0.7068966 0.6844584 0.5500873 351 51 749 123
145 0.025 0.45 0.7183908 0.6891680 0.4189891 347 49 753 125
146 0.025 0.46 0.7298851 0.6915228 0.2753680 346 47 754 127
147 0.025 0.47 0.7356322 0.7009419 0.3238852 335 46 765 128
148 0.025 0.48 0.7413793 0.7040816 0.2843481 332 45 768 129
149 0.025 0.49 0.7471264 0.7072214 0.2479759 329 44 771 130
150 0.025 0.50 0.7471264 0.7158556 0.3714097 318 44 782 130
151 0.025 0.51 0.7586207 0.7213501 0.2760944 313 42 787 132
152 0.025 0.52 0.7643678 0.7299843 0.3136785 303 41 797 133
153 0.025 0.53 0.7643678 0.7339089 0.3755397 298 41 802 133
154 0.025 0.54 0.7758621 0.7448980 0.3603058 286 39 814 135
155 0.025 0.55 0.7758621 0.7503925 0.4585858 279 39 821 135
156 0.025 0.56 0.7758621 0.7574568 0.6069666 270 39 830 135
157 0.025 0.57 0.7816092 0.7653061 0.6528283 261 38 839 136
158 0.025 0.58 0.7816092 0.7692308 0.7487775 256 38 844 136
159 0.025 0.59 0.7873563 0.7731554 0.7010032 252 37 848 137
160 0.025 0.60 0.7931034 0.7802198 0.7314768 244 36 856 138
161 0.025 0.61 0.7931034 0.7841444 0.8336838 239 36 861 138
162 0.025 0.62 0.7931034 0.7849294 0.8546904 238 36 862 138
163 0.025 0.63 0.7988506 0.7888540 0.8043177 234 35 866 139
164 0.025 0.64 0.8045977 0.7967033 0.8594275 225 34 875 140
165 0.025 0.65 0.8218391 0.8061224 0.6446717 216 31 884 143
166 0.025 0.66 0.8218391 0.8092622 0.7258825 212 31 888 143
167 0.025 0.67 0.8275862 0.8186813 0.8241371 201 30 899 144
168 0.025 0.68 0.8390805 0.8281005 0.7603588 191 28 909 146
169 0.025 0.69 0.8505747 0.8351648 0.6314783 184 26 916 148
170 0.025 0.70 0.8505747 0.8367347 0.6736090 182 26 918 148
171 0.025 0.71 0.8505747 0.8414443 0.8078706 176 26 924 148
172 0.025 0.72 0.8620690 0.8485086 0.6721908 169 24 931 150
173 0.025 0.73 0.8620690 0.8547881 0.8590482 161 24 939 150
174 0.025 0.74 0.8678161 0.8594976 0.8239815 156 23 944 151
175 0.025 0.75 0.8735632 0.8634223 0.7638518 152 22 948 152
176 0.025 0.76 0.8793103 0.8681319 0.7275054 147 21 953 153
177 0.025 0.77 0.8850575 0.8744113 0.7391619 140 20 960 154
178 0.025 0.78 0.8850575 0.8806907 0.9478620 132 20 968 154
179 0.025 0.79 0.8850575 0.8822606 1.0000000 130 20 970 154
180 0.025 0.80 0.8965517 0.8893250 0.8438452 123 18 977 156
181 0.025 0.81 0.9022989 0.8924647 0.7497552 120 17 980 157
182 0.025 0.82 0.9022989 0.8956044 0.8592023 116 17 984 157
183 0.025 0.83 0.9022989 0.9018838 1.0000000 108 17 992 157
184 0.025 0.84 0.9022989 0.9089482 0.8521950 99 17 1001 157
185 0.025 0.85 0.9195402 0.9152276 0.9415352 94 14 1006 160
186 0.025 0.86 0.9252874 0.9207221 0.9291722 88 13 1012 161
187 0.025 0.87 0.9252874 0.9301413 0.9121818 76 13 1024 161
188 0.025 0.88 0.9425287 0.9356358 0.8161399 72 10 1028 164
189 0.025 0.89 0.9425287 0.9379906 0.9219472 69 10 1031 164
190 0.025 0.90 0.9597701 0.9442700 0.4345871 64 7 1036 167
191 0.025 0.91 0.9597701 0.9481947 0.5772773 59 7 1041 167
192 0.025 0.92 0.9597701 0.9521193 0.7507696 54 7 1046 167
193 0.025 0.93 0.9655172 0.9591837 0.8039487 46 6 1054 168
194 0.025 0.94 0.9770115 0.9686028 0.6522932 36 4 1064 170
195 0.025 0.95 0.9827586 0.9740973 0.6049841 30 3 1070 171
196 0.025 0.96 0.9827586 0.9819466 1.0000000 20 3 1080 171
197 0.025 0.97 0.9827586 0.9866562 0.8991846 14 3 1086 171
198 0.025 0.98 0.9942529 0.9937206 1.0000000 7 1 1093 173
199 0.025 0.99 1.0000000 0.9968603 0.9461552 4 0 1096 174
200 0.025 1.00 1.0000000 1.0000000 0.0000000 0 0 1100 174#VCs appear to be better on both, but we'll see in final combining of it--pretty sure it's the second set of doing it that's wthe way I did it for HOMER, but double-check.Now that I’ve seen some enrichment of differential contact (DC) in differential expression (DE) based on my linear modeling results from before, I would like to further quantify this effect. In order to do so, I now extract log2 observed/expected homer-normalized contact frequency values and RPKM expression values for each gene/bin set, so I can look at correlations of these values and assess their explanatory power.
Contact Frequency Extraction
In this section, I proceed to create a function in order to extract the Hi-C interaction frequency values for the different types of summaries I’ve made gene overlaps with above. This allows for operations to be performed separately utilizing different summaries of Hi-C contacts.
######Get a df with the H and C coordinates of the hits, and the IF values from homer. This subset df makes things easier to extract. Both for VC and KR.
###VC
contacts.VC <- data.frame(h1=data.VC$H1, h2=data.VC$H2, c1=data.VC$C1, c2=data.VC$C2, A_21792_HIC=data.VC$`A-21792_VC`, B_28126_HIC=data.VC$`B-28126_VC`, C_3649_HIC=data.VC$`C-3649_VC`, D_40300_HIC=data.VC$`D-40300_VC`, E_28815_HIC=data.VC$`E-28815_VC`, F_28834_HIC=data.VC$`F-28834_VC`, G_3624_HIC=data.VC$`G-3624_VC`, H_3651_HIC=data.VC$`H-3651_VC`, stringsAsFactors = FALSE)
newH1 <- as.numeric(gsub(".*-", "", contacts.VC$h1))
newH2 <- as.numeric(gsub(".*-", "", contacts.VC$h2))
lower.HID <- ifelse(newH1<newH2, contacts.VC$h1, contacts.VC$h2)
higher.HID2 <- ifelse(newH1<newH2, contacts.VC$h2, contacts.VC$h1)
contacts.VC$hpair <- paste(lower.HID, higher.HID2, sep="_")
newC1 <- as.numeric(gsub(".*-", "", contacts.VC$c1))
newC2 <- as.numeric(gsub(".*-", "", contacts.VC$c2))
lower.CID <- ifelse(newC1<newC2, contacts.VC$c1, contacts.VC$c2)
higher.CID2 <- ifelse(newC1<newC2, contacts.VC$c2, contacts.VC$c1)
contacts.VC$cpair <- paste(lower.CID, higher.CID2, sep="_")
###KR
contacts.KR <- data.frame(h1=data.KR$H1, h2=data.KR$H2, c1=data.KR$C1, c2=data.KR$C2, A_21792_HIC=data.KR$`A-21792_KR`, B_28126_HIC=data.KR$`B-28126_KR`, C_3649_HIC=data.KR$`C-3649_KR`, D_40300_HIC=data.KR$`D-40300_KR`, E_28815_HIC=data.KR$`E-28815_KR`, F_28834_HIC=data.KR$`F-28834_KR`, G_3624_HIC=data.KR$`G-3624_KR`, H_3651_HIC=data.KR$`H-3651_KR`, stringsAsFactors = FALSE)
newH1 <- as.numeric(gsub(".*-", "", contacts.KR$h1))
newH2 <- as.numeric(gsub(".*-", "", contacts.KR$h2))
lower.HID <- ifelse(newH1<newH2, contacts.KR$h1, contacts.KR$h2)
higher.HID2 <- ifelse(newH1<newH2, contacts.KR$h2, contacts.KR$h1)
contacts.KR$hpair <- paste(lower.HID, higher.HID2, sep="_")
newC1 <- as.numeric(gsub(".*-", "", contacts.KR$c1))
newC2 <- as.numeric(gsub(".*-", "", contacts.KR$c2))
lower.CID <- ifelse(newC1<newC2, contacts.KR$c1, contacts.KR$c2)
higher.CID2 <- ifelse(newC1<newC2, contacts.KR$c2, contacts.KR$c1)
contacts.KR$cpair <- paste(lower.CID, higher.CID2, sep="_")
#Fix column names for both contacts.VC and contacts.KR
colnames(contacts.KR)[5:12] <- colnames(contacts.VC)[5:12] <- c("A_21792_HIC", "B_28126_HIC", "C_3649_HIC", "D_40300_HIC", "E_28815_HIC", "F_28834_HIC", "G_3624_HIC", "H_3651_HIC")
#A function that takes a dataframe (like gene.hic.filt) and two columns from the dataframe to create a pair vector for the given interaction. First ensures the first bin in a pair is always lowest to make this easier. Then extracts the IF values for that vector from the contacts df created above. This provides me with the appropriate Hi-C data values for the different bin classes we're examining here, so that I can later test them with linear modeling to quantify their effect on expression.
IF.extractor <- function(dataframe, col1, col2, contacts, species, strand=FALSE){
new1 <- as.numeric(gsub(".*-", "", dataframe[,col1]))
if(strand==FALSE){#In the case where I'm not worried about strand, I just work with the second column selected.
new2 <- as.numeric(gsub(".*-", "", dataframe[,col2]))
lower1 <- ifelse(new1<new2, dataframe[,col1], dataframe[,col2])
higher2 <- ifelse(new1<new2, dataframe[,col2], dataframe[,col1]) #Fix all the columns first
if(species=="H"){ #Then depending on species create the pair column and merge to contact info.
dataframe[,"hpair"] <- paste(lower1, higher2, sep="_")
finaldf <- left_join(dataframe, contacts[,c("A_21792_HIC", "B_28126_HIC", "C_3649_HIC", "D_40300_HIC", "E_28815_HIC", "F_28834_HIC", "G_3624_HIC", "H_3651_HIC", "hpair")], by="hpair")
}
else if(species=="C"){
dataframe[,"cpair"] <- paste(lower1, higher2, sep="_")
finaldf <- left_join(dataframe, contacts[,c("A_21792_HIC", "B_28126_HIC", "C_3649_HIC", "D_40300_HIC", "E_28815_HIC", "F_28834_HIC", "G_3624_HIC", "H_3651_HIC", "cpair")], by="cpair")
}
}
else if(strand==TRUE){#If dealing with upstream and downstream hits, need to do things separately for genes on the + and - strand.
if(species=="H"){
US <- ifelse(dataframe[,"Hstrand.H"]=="+", dataframe[,"US_bin.H"], dataframe[,"DS_bin.H"]) #Obtain upstream bins depending on strand.
new2 <- as.numeric(gsub(".*-", "", US)) #Now rearrange the pairs to ensure regardless of stream we can find the pair (first mate lower coordinates than 2nd).
lower1 <- ifelse(new1<new2, dataframe[,col1], US)
higher2 <- ifelse(new1<new2, US, dataframe[,col1])
dataframe[,"hpair"] <- paste(lower1, higher2, sep="_")
dataframe[,"USFDR"] <- ifelse(dataframe[,"Hstrand.H"]=="+", dataframe[,"US_FDR.H"], dataframe[,"DS_FDR.H"])
finaldf <- left_join(dataframe, contacts[,c("A_21792_HIC", "B_28126_HIC", "C_3649_HIC", "D_40300_HIC", "E_28815_HIC", "F_28834_HIC", "G_3624_HIC", "H_3651_HIC", "hpair")], by="hpair")
}
else if(species=="C"){
US <- ifelse(dataframe[,"Hstrand.C"]=="+", dataframe[,"US_bin.C"], dataframe[,"DS_bin.C"]) #Obtain upstream bins depending on strand.
new2 <- as.numeric(gsub(".*-", "", US)) #Now rearrange the pairs to ensure regardless of stream we can find the pair (first mate lower coordinates than 2nd).
lower1 <- ifelse(new1<new2, dataframe[,col1], US)
higher2 <- ifelse(new1<new2, US, dataframe[,col1])
dataframe[,"cpair"] <- paste(lower1, higher2, sep="_")
dataframe[,"USFDR"] <- ifelse(dataframe[,"Hstrand.C"]=="+", dataframe[,"US_FDR.C"], dataframe[,"DS_FDR.C"])
finaldf <- left_join(dataframe, contacts[,c("A_21792_HIC", "B_28126_HIC", "C_3649_HIC", "D_40300_HIC", "E_28815_HIC", "F_28834_HIC", "G_3624_HIC", "H_3651_HIC", "cpair")], by="cpair")
}
}
#before finally returning, remove rows where we don't have full Hi-C data.
finaldf <- finaldf[complete.cases(finaldf[,c("A_21792_HIC", "B_28126_HIC", "C_3649_HIC", "D_40300_HIC", "E_28815_HIC", "F_28834_HIC", "G_3624_HIC", "H_3651_HIC")]),]
return(finaldf)
}
#Now I can use the IF.extractor function to make a number of different dataframes for actually testing different IF values with the RPKM expression values.
h_minFDR.KR <- IF.extractor(gene.hic.KR, "HID", "min_FDR_bin.H", contacts.KR, "H")
h_minFDR.VC <- IF.extractor(gene.hic.VC, "HID", "min_FDR_bin.H", contacts.VC, "H")
#h_maxB <- IF.extractor(gene.hic.filt, "HID", "max_B_bin.H", contacts, "H")
#c_maxB <- IF.extractor(gene.hic.filt, "CID", "max_B_bin.C", contacts, "C")
#h_US <- IF.extractor(gene.hic.filt, "HID", "US_bin.H", contacts, "H", TRUE)
#c_US <- IF.extractor(gene.hic.filt, "CID", "US_bin.C", contacts, "C", TRUE)
#Write these out so they can be permuted upon on midway2.
fwrite(h_minFDR.KR, "~/Desktop/Hi-C/HiC_covs/juicer/h_minFDR.KR", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)
fwrite(h_minFDR.VC, "~/Desktop/Hi-C/HiC_covs/juicer/h_minFDR.VC", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)
#fwrite(h_maxB, "~/Desktop/Hi-C/HiC_covs/h_maxB", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)
#fwrite(c_maxB, "~/Desktop/Hi-C/HiC_covs/c_maxB", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)
#fwrite(h_US, "~/Desktop/Hi-C/HiC_covs/h_US", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)
#fwrite(c_US, "~/Desktop/Hi-C/HiC_covs/c_US", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)
#Now the same thing, but subsetting down to ONLY the genes that show evidence for DE at 5% FDR.
h_minFDR_DE.KR <- filter(h_minFDR.KR, adj.P.Val<=0.05)
h_minFDR_DE.VC <- filter(h_minFDR.VC, adj.P.Val<=0.05)
#h_maxB_DE <- filter(h_maxB, adj.P.Val<=0.05)
#c_maxB_DE <- filter(c_maxB, adj.P.Val<=0.05)
#h_US_DE <- filter(h_US, adj.P.Val<=0.05)
#c_US_DE <- filter(c_US, adj.P.Val<=0.05)
fwrite(h_minFDR_DE.KR, "~/Desktop/Hi-C/HiC_covs/juicer/h_minFDR_DE.KR", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)
fwrite(h_minFDR_DE.VC, "~/Desktop/Hi-C/HiC_covs/juicer/h_minFDR_DE.VC", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)
#fwrite(h_maxB_DE, "~/Desktop/Hi-C/HiC_covs/h_maxB_DE", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)
#fwrite(c_maxB_DE, "~/Desktop/Hi-C/HiC_covs/c_maxB_DE", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)
#fwrite(h_US_DE, "~/Desktop/Hi-C/HiC_covs/h_US_DE", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)
#fwrite(c_US_DE, "~/Desktop/Hi-C/HiC_covs/c_US_DE", quote = FALSE, sep = "\t", na = "NA", col.names = TRUE)Now I use these data frames and join each again on the RPKM table by genes, in order to obtain RPKM expression values for each gene-Hi-C bin set.
Merging contact frequency values with expression values.
In this next section I join each of the data frames created above on the RPKM table by genes, in order to have a merged table with expression values and contact frequencies for each gene-Hi-C bin set. I move then to explore correlations between the values.
#Join all of the previously-made Hi-C interaction frequency tables to the RPKM table by genes, to obtain RPKM values in concert with contact frequency values.
RPKM <- as.data.frame(weighted.data$E)
RPKM$genes <- rownames(RPKM)
hmin.KR <- left_join(h_minFDR.KR, RPKM, by="genes")
hmin.VC <- left_join(h_minFDR.VC, RPKM, by="genes")
#hmaxB <- left_join(h_maxB, RPKM, by="genes")
#cmaxB <- left_join(c_maxB, RPKM, by="genes")
#hUS <- left_join(h_US, RPKM, by="genes")
#cUS <- left_join(c_US, RPKM, by="genes")
#Get the same thing filtered for only DE genes.
hminDE.KR <- filter(hmin.KR, adj.P.Val<=0.05)
hminDE.VC <- filter(hmin.VC, adj.P.Val<=0.05)
#hmaxBDE <- filter(hmaxB, adj.P.Val<=0.05)
#cmaxBDE <- filter(cmaxB, adj.P.Val <=0.05)
#hUSDE <- filter(hUS, adj.P.Val<=0.05)
#cUSDE <- filter(cUS, adj.P.Val<=0.05)
hmin_noDE.KR <- filter(hmin.KR, adj.P.Val>0.05) #Pull out specific set of non-DE hits.
hmin_noDE.VC <- filter(hmin.VC, adj.P.Val>0.05)
#Extract contacts and expression for the contact with the lowest FDR from linear modeling.
KRcontacts <- hmin.KR[,61:68]
KRexprs <- hmin.KR[,69:76]
KRcontactsDE <- hminDE.KR[,61:68]
KRexprsDE <- hminDE.KR[,69:76]
nonDEcontactsKR <- hmin_noDE.KR[,61:68]
nonDEexprsKR <- hmin_noDE.KR[,69:76]
VCcontacts <- hmin.VC[,61:68]
VCexprs <- hmin.VC[,69:76]
VCcontactsDE <- hminDE.VC[,61:68]
VCexprsDE <- hminDE.VC[,69:76]
nonDEcontactsVC <- hmin_noDE.VC[,61:68]
nonDEexprsVC <- hmin_noDE.VC[,69:76]
#Function for calculating gene-wise correlations.
cor.calc <- function(hicdata, exprdata){
cor.exp <- vector(length=nrow(hicdata))
for(i in 1:nrow(hicdata)){
cor.exp[i] <- cor(as.numeric(hicdata[i,]), as.numeric(exprdata[i,]))
}
return(cor.exp)
}
#Calculate correlations for different sets! I've commented out the species-specific calculations here because they just aren't that interesting. If you look at DE dynamics within each species you do NOT get a gorgeous bimodal as you do for across--looks more uniform and messy.
fullcorsKR <- data.frame(cor=cor.calc(KRcontacts, KRexprs), type="all")
fullDEcorsKR <- data.frame(cor=cor.calc(KRcontactsDE, KRexprsDE), type="DE")
fullnoDEcorsKR <- data.frame(cor=cor.calc(nonDEcontactsKR, nonDEexprsKR), type="non-DE")
#VC now
fullcorsVC <- data.frame(cor=cor.calc(VCcontacts, VCexprs), type="all")
fullDEcorsVC <- data.frame(cor=cor.calc(VCcontactsDE, VCexprsDE), type="DE")
fullnoDEcorsVC <- data.frame(cor=cor.calc(nonDEcontactsVC, nonDEexprsVC), type="non-DE")
#Combine these dfs to plot in one gorgeous ggplot!
ggcorsKR <- rbind(fullcorsKR, fullDEcorsKR, fullnoDEcorsKR)
ggcorsVC <- rbind(fullcorsVC, fullDEcorsVC, fullnoDEcorsVC)
#VC then KR
ggplot(data=filter(ggcorsVC, type=="all"|type=="DE"|type=="non-DE")) + stat_density(aes(x=cor, group=type, color=type, y=..scaled..), position="identity", geom="line") + ggtitle("Correlation b/t RPKM Expression and VC Hi-C Contact Frequency") + xlab("Pearson Correlations b/t RPKM Expression and VC Hi-C Contact Frequency") + ylab("Density") + scale_color_manual("Gene Set", values=c("red", "blue", "green"), labels=c("All genes", "DE genes", "non-DE genes"))
ggplot(data=filter(ggcorsKR, type=="all"|type=="DE"|type=="non-DE")) + stat_density(aes(x=cor, group=type, color=type, y=..scaled..), position="identity", geom="line") + ggtitle("Correlation b/t RPKM Expression and KR Hi-C Contact Frequency") + xlab("Pearson Correlations b/t RPKM Expression and KR Hi-C Contact Frequency") + ylab("Density") + scale_color_manual("Gene Set", values=c("red", "blue", "green"), labels=c("All genes", "DE genes", "non-DE genes"))
###This looks great, showing strong bimodal distribution for the DE genes and broader distributions with a peak at 0 for the non-DE set and the full set. To assess if this is legit at all, I now re-run this correlation analysis after permuting the Hi-C values. I shuffle sample IDs on a gene-by-gene basis to accomplish this:
cor.permuter <- function(hicdata, exprdata, nperm){
result <- data.frame(cor=NA, type=rep(1:nperm, each=nrow(exprdata)))
for(perm in 1:nperm){
permute <- hicdata
for(row in 1:nrow(hicdata)){
permute[row,] <- sample(hicdata[row,])
}
myindices <- which(result$type==perm)
result[myindices,1] <- cor.calc(permute, exprdata)
}
return(result)
}
#Just do it with 5 permutations to see the general effect quickly:
full.perm.KR <- cor.permuter(KRcontacts, KRexprs, 5)
DE.perm.KR <- cor.permuter(KRcontactsDE, KRexprsDE, 5)
nonDE.perm.KR <- cor.permuter(nonDEcontactsKR, nonDEexprsKR, 5)
full.perm.VC <- cor.permuter(VCcontacts, VCexprs, 5)
DE.perm.VC <- cor.permuter(VCcontactsDE, VCexprsDE, 5)
nonDE.perm.VC <- cor.permuter(nonDEcontactsVC, nonDEexprsVC, 5)
#Now visualize.
ggplot(data=filter(ggcorsKR, type=="all"|type=="DE"|type=="non-DE")) + stat_density(aes(x=cor, group=type, color=type, y=..scaled..), position="identity", geom="line") + stat_density(data=full.perm.KR, aes(x=cor, group=type), geom="line", linetype="dotted", position="identity") + stat_density(data=DE.perm.KR, aes(x=cor, group=type), geom="line", linetype="dashed", position="identity") + stat_density(data=nonDE.perm.KR, aes(x=cor, group=type), geom="line", linetype="twodash", position="identity") + ggtitle("Correlation b/t RPKM Expression and KR Hi-C Contact Frequency") + xlab("Pearson Correlations b/t RPKM Expression and KR Hi-C Contact Frequency") + ylab("Density") + scale_color_manual("Gene Set", values=c("red", "blue", "green"), labels=c("All genes", "DE genes", "non-DE genes"))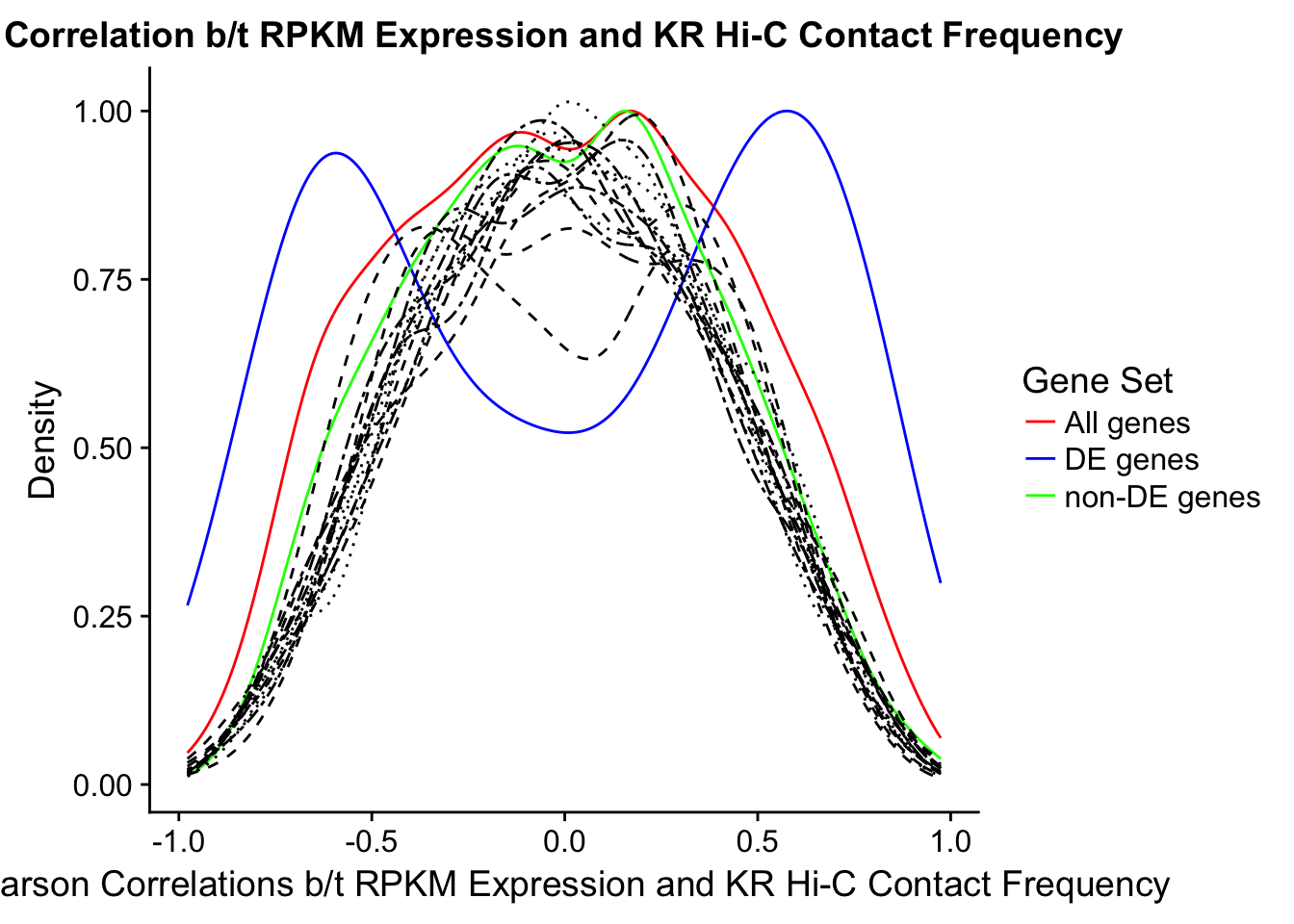
ggplot(data=filter(ggcorsVC, type=="all"|type=="DE"|type=="non-DE")) + stat_density(aes(x=cor, group=type, color=type, y=..scaled..), position="identity", geom="line") + stat_density(data=full.perm.VC, aes(x=cor, group=type), geom="line", linetype="dotted", position="identity") + stat_density(data=DE.perm.VC, aes(x=cor, group=type), geom="line", linetype="dashed", position="identity") + stat_density(data=nonDE.perm.VC, aes(x=cor, group=type), geom="line", linetype="twodash", position="identity") + ggtitle("Correlation b/t RPKM Expression and VC Hi-C Contact Frequency") + xlab("Pearson Correlations b/t RPKM Expression and VC Hi-C Contact Frequency") + ylab("Density") + scale_color_manual("Gene Set", values=c("red", "blue", "green"), labels=c("All genes", "DE genes", "non-DE genes"))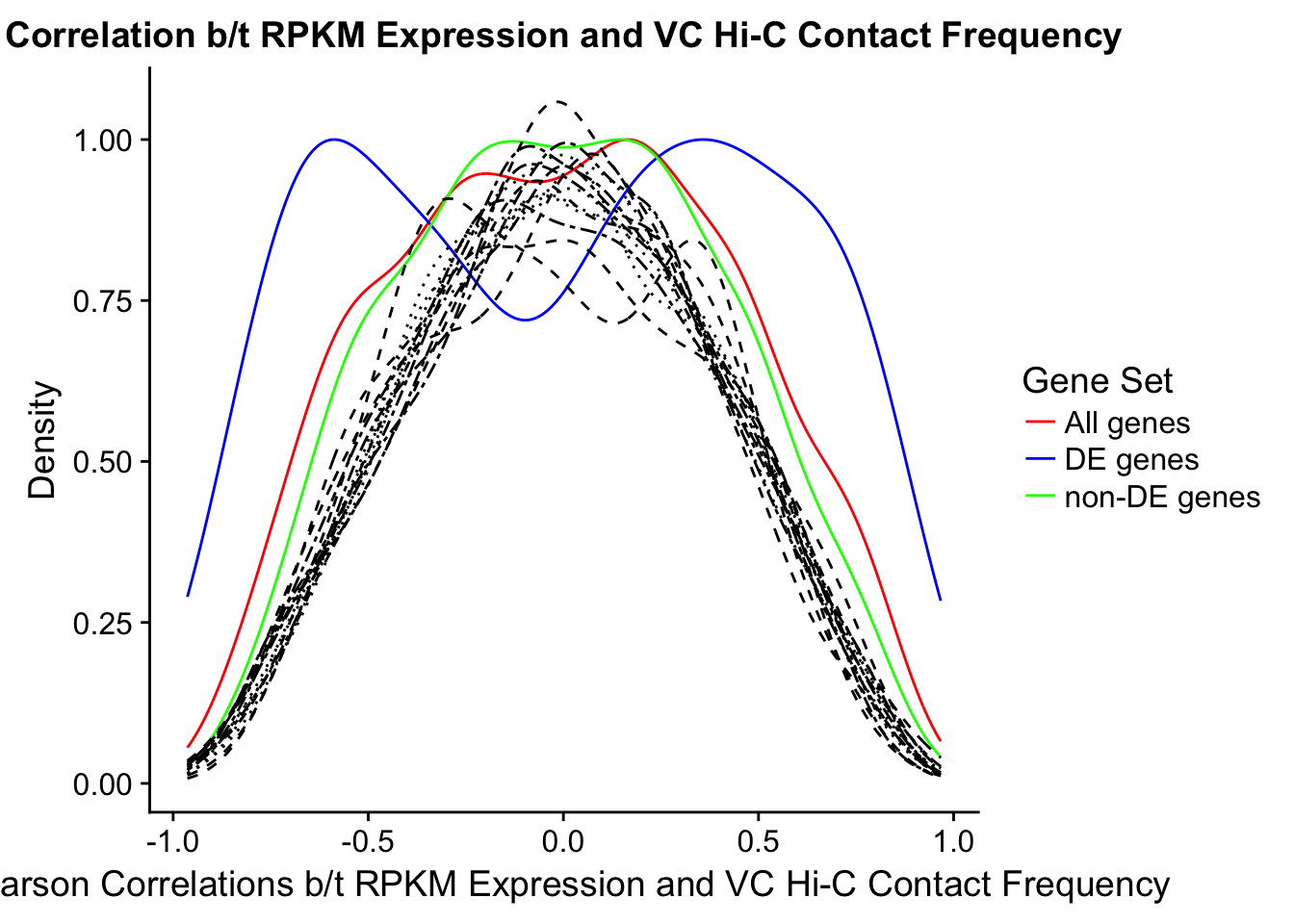
#See that permuted datasets have a tighter correlation distribution with a strong peak at 0. Reassuring. In the future I will repeat this analysis on Midway2, running 10000 permutations to see the full range of permuted data possible.Now that I’ve seen some nice effects in the correlation between expression and contact for different sets of genes, and for permutations on those sets, I move to see what kind of quantitiative explanatory power differential contact (DC) might actually have for differential expression (DE).
How well does Hi-C data explain expression data?
In this next section I “regress out” the effect of Hi-C contacts from their overlapping genes’ RPKM expression values, comparing a linear model run on the base values to one run on the residuals of expression after regressing out Hi-C data. Comparing the p-values before and after this regression can give some sense of whether the DE is being driven by differential Hi-C contacts (DC).
###A function to calculate gene-wise correlations between Hi-C data and expression data, but for spearman correlations. Pearson correlation calculator function is in the chunk above.
cor.calc.spear <- function(hicdata, exprdata){
cor.exp <- vector(length=nrow(hicdata))
for(i in 1:nrow(hicdata)){
cor.exp[i] <- cor(as.numeric(hicdata[i,]), as.numeric(exprdata[i,]), method="spearman")
}
return(cor.exp)
}
###A function to permute a Hi-C df by going gene-by-gene (row-by-row) and shuffling all sample IDs.
shuffler <- function(hicdata){
for(row in 1:nrow(hicdata)){
hicdata[row,] <- sample(hicdata[row,])
}
return(hicdata)
}
###A function that runs a linear model, both with and without Hi-C corrected expression values, and returns a dataframe of hit classes (DE or not before and after correction). Also spits back out p-values before and after correction, as well as correlations between Hi-C data and expression data. Since the former is a one-row data frame of 4 points and the latter is a 3-column data frame with the number of genes rows, returns a list.
lmcorrect <- function(voom.obj, exprs, cov_matrix, meta_df){
mygenes <- cov_matrix$genes #Pull out the relevant genes here; used for subsetting the voom.obj in a bit.
cov_matrix <- cov_matrix[, -9] #Remove genes from the cov matrix
hic_present <- sapply(1:nrow(cov_matrix), function(i) !any(is.na(cov_matrix[i,]))) #First, remove any rows w/ missing Hi-C data.
exprs <- data.matrix(exprs[hic_present,]) #Filter expression with this
cov_matrix <- data.matrix(cov_matrix[hic_present,]) #Filter Hi-C data with this
#Now, prepare to run the actual models. First run a model w/ Hi-C as a covariate to evaluate
SP <- factor(meta_df$SP,levels = c("H","C"))
design <- model.matrix(~0+SP)
colnames(design) <- c("Human", "Chimp")
resid_hic <- array(0, dim=c(nrow(exprs), ncol(cov_matrix))) #Initialize a dataframe for storing the residuals.
for(i in 1:nrow(exprs)){#Loop through rows of the expression df, running linear modeling w/ Hi-C to obtain residuals.
resid_hic[i,] <- lm(exprs[i,]~cov_matrix[i,])$resid
}
mycon <- makeContrasts(HvC = Human-Chimp, levels = design)
#Filter the voom object to only contain genes that had Hi-C information here.
good.indices <- which(rownames(voom.obj$E) %in% mygenes)
voom.obj <- voom.obj[good.indices,]
#Now, replace the RPKM values in the voom object for after linear modeling with the residuals.
voom.obj.after <- voom.obj
voom.obj.after$E <- resid_hic
lmFit(voom.obj, design=design) %>% eBayes(.) %>% contrasts.fit(., mycon) %>% eBayes(.) %>% topTable(., coef = 1, adjust.method = "BH", number = Inf, sort.by="none") -> fit_before
lmFit(voom.obj.after, design=design) %>% eBayes(.) %>% contrasts.fit(., mycon) %>% eBayes(.) %>% topTable(., coef = 1, adjust.method = "BH", number = Inf, sort.by="none") -> fit_after
result.DE.cats <- data.frame(DEneither=sum(fit_before$adj.P.Val>0.05&fit_after$adj.P.Val>0.05), DEbefore=sum(fit_before$adj.P.Val<=0.05&fit_after$adj.P.Val>0.05), DEafter=sum(fit_before$adj.P.Val>0.05&fit_after$adj.P.Val<=0.05), DEboth=sum(fit_before$adj.P.Val<=0.05&fit_after$adj.P.Val<=0.05))
result.DE.stats <- data.frame(cor.pear=cor.calc(cov_matrix, exprs), cor.spear=cor.calc.spear(cov_matrix, exprs), pval.before=fit_before$adj.P.Val, pval.after=fit_after$adj.P.Val)
result <- list("categories"=result.DE.cats, "stats"=result.DE.stats)
return(result)
}
#Proceed to use the function on the different dataframes I've created with different sets of overlapping Hi-C contacts:
h_minFDR_pvals_KR <- lmcorrect(weighted.data, hmin.KR[,c("A-21792", "B-28126", "C-3649", "D-40300", "E-28815", "F-28834", "G-3624", "H-3651")], hmin.KR[,c("A_21792_HIC", "B_28126_HIC", "C_3649_HIC", "D_40300_HIC", "E_28815_HIC", "F_28834_HIC", "G_3624_HIC", "H_3651_HIC", "genes")], meta.data)
h_minFDR_pvals_VC <- lmcorrect(weighted.data, hmin.VC[,c("A-21792", "B-28126", "C-3649", "D-40300", "E-28815", "F-28834", "G-3624", "H-3651")], hmin.VC[,c("A_21792_HIC", "B_28126_HIC", "C_3649_HIC", "D_40300_HIC", "E_28815_HIC", "F_28834_HIC", "G_3624_HIC", "H_3651_HIC", "genes")], meta.data)
#I now proceed to visualize the difference in p-values for expression before and after "correcting" for the Hi-C data. I am hoping to see many hits in the bottom right quadrant of the following plots, indicating genes that showed up as DE before Hi-C correction, but not after. I also expect that p-values falling farther away from the null line of expectation for p-values being identical between models will have higher correlations, which I color here for the pearson correlation (results look similar for spearman).
#ggplot(data=h_minFDR_pvals_KR$stats, aes(x=-log10(pval.before), y=-log10(pval.after), color=abs(cor.pear))) + geom_point(size=0.01) + geom_hline(yintercept=-log10(0.05), color="red") + geom_vline(xintercept=-log10(0.05), color="red") + geom_abline(slope=1, intercept=0, color="green", linetype="dashed") + ggtitle("Evidence for DE before vs. after regressing out Hi-C min. FDR, KR") + xlab("-log10(p-value of DE before Hi-C regression)") + ylab("-log10(p-value of DE after Hi-C regression")
ggplot(data=h_minFDR_pvals_KR$stats, aes(x=-log10(pval.before), y=-log10(pval.after))) + geom_point(size=0.01) + geom_hline(yintercept=-log10(0.05), color="red") + geom_vline(xintercept=-log10(0.05), color="red") + geom_abline(slope=1, intercept=0, color="green", linetype="dashed") + ggtitle("Evidence for DE Before vs. After Regressing out Hi-C Contact Frequency") + xlab("-log10(p-value of DE before KR Hi-C regression)") + ylab("-log10(p-value of DE after KR Hi-C regression)") + theme(plot.title=element_text(hjust=1))ggplot(data=h_minFDR_pvals_VC$stats, aes(x=-log10(pval.before), y=-log10(pval.after))) + geom_point(size=0.01) + geom_hline(yintercept=-log10(0.05), color="red") + geom_vline(xintercept=-log10(0.05), color="red") + geom_abline(slope=1, intercept=0, color="green", linetype="dashed") + ggtitle("Evidence for DE Before vs. After Regressing out Hi-C Contact Frequency") + xlab("-log10(p-value of DE before VC Hi-C regression)") + ylab("-log10(p-value of DE after VC Hi-C regression)") + theme(plot.title=element_text(hjust=1)) Critically, here I see that “regressing out” Hi-C data and trying to model expression again moves many genes from being significantly differentially expressed (at FDR of 5%) to no longer showing differential expression. Seeing most of the hits in the bottom right corner of these visualizations is what confirms this. Reassuringly, I also see stronger correlations between RPKM expression values and normalized Hi-C interaction frequency values for points farther away from the diagonal green line of expectation. However, since this is not a statistical test, I have no assessment of significance. To accomplish this, I compare these results to running the same kind of analysis on permuted data in the next section.
Differences in effect size/sign changes
This next section would be to look at differences in effect size and sign changes when doing the two separate lms (one before and one after regressing out Hi-C data from RPKM expression). Unfortunately the ash results I get have all their svalues as 1 so this is boring/not interesting right now. Hence it is not included. But let’s give it another go!
#######FRESH CODE 6-21-18#########
design <- model.matrix(~meta.data$SP)
exprs <- select(hmin.KR, "A-21792", "B-28126", "C-3649", "D-40300", "E-28815", "F-28834", "G-3624", "H-3651", genes) #Should be done on just DE genes, filter with adj.P.Val<=0.05
hic <- select(hmin.KR,"A_21792_HIC", "B_28126_HIC", "C_3649_HIC", "D_40300_HIC", "E_28815_HIC", "F_28834_HIC", "G_3624_HIC", "H_3651_HIC")
present <- sapply(1:nrow(hic), function(i) !any(is.na(hic[i,])))
genes <- exprs$genes
DEgenes <- filter(hmin.KR, adj.P.Val<=0.05) %>% select(., genes)
myDEindices <- which(genes %in% DEgenes[,1])
exprs <- exprs[,-9]
exprs <- data.matrix(exprs[present,])
hic <- data.matrix(hic[present,])
designy <- as.factor(c("human", "human", "chimp", "chimp", "human", "human", "chimp", "chimp"))
colnames(hic) <- colnames(exprs)
hic <- as.data.table(hic)
exprs <- as.data.table(exprs)
test <- mediate.test.regressing(exprs, designy, hic)
Attaching package: 'assertthat'The following object is masked from 'package:tibble':
has_namevshrDE <- vash(test$d_se[myDEindices], df=6)
vshrnonDE <- vash(test$d_se[-myDEindices], df=6)
vshrink <- vash(test$d_se, df=6)
DEtest <- test[myDEindices,]
nonDEtest <- test[-myDEindices,]
ash_reg <- ash(as.vector(DEtest$d), as.vector(vshrDE$sd.post), mixcompdist = "normal") #Now it works with this design coding. Use 6 df, as 2-group comparison and 8 individual samples (samples - 2)Due to absence of package REBayes, switching to EM algorithmsummary(ash_reg$result$svalue<=0.05) #Incredible result, looks too good to be true. Mode FALSE TRUE
logical 245 18 ind_sig <- (ash_reg$result$svalue < 0.05)
ind_sig[ind_sig == TRUE] <- "red"
ind_sig[ind_sig == FALSE] <- "black"
plot(x=ash_reg$result$betahat, y=ash_reg$result$sebetahat, ylab="Standard error of the difference", xlab="Difference in effect size from 2 lms, KR", main="Difference of effect size (DE genes)", pch=16, cex=0.6, col=ind_sig)#, xlim=c(-2.5, 2.5), ylim=c(0, 1.3))
legend("topleft", legend=c("s < 0.05", "s >= 0.05"), col=c("red", "black"), pch=16:16, cex=0.8)
ash_reg_nonDE <- ash(as.vector(nonDEtest$d), as.vector(vshrnonDE$sd.post), mixcompdist = "normal") #Now it works with this design coding. Use 6 df, as 2-group comparison and 8 individual samples (samples - 2)Due to absence of package REBayes, switching to EM algorithmsummary(ash_reg_nonDE$result$svalue<=0.05) #Incredible result, looks too good to be true. Mode FALSE
logical 1011 ind_sig <- (ash_reg_nonDE$result$svalue < 0.05)
ind_sig[ind_sig == TRUE] <- "red"
ind_sig[ind_sig == FALSE] <- "black"
plot(x=ash_reg_nonDE$result$betahat, y=ash_reg_nonDE$result$sebetahat, ylab="Standard error of the difference", xlab="Difference in effect size from 2 lms, KR", main="Difference of effect size (non-DE genes)", pch=16, cex=0.6, col=ind_sig)#, xlim=c(-2.5, 2.5), ylim=c(0, 1.3))
legend("topleft", legend=c("s < 0.05", "s >= 0.05"), col=c("red", "black"), pch=16:16, cex=0.8)
exprs <- select(hmin.VC, "A-21792", "B-28126", "C-3649", "D-40300", "E-28815", "F-28834", "G-3624", "H-3651", genes) #Should be done on just DE genes, filter with adj.P.Val<=0.05
hic <- select(hmin.VC,"A_21792_HIC", "B_28126_HIC", "C_3649_HIC", "D_40300_HIC", "E_28815_HIC", "F_28834_HIC", "G_3624_HIC", "H_3651_HIC")
present <- sapply(1:nrow(hic), function(i) !any(is.na(hic[i,])))
genes <- exprs$genes
DEgenes <- filter(hmin.KR, adj.P.Val<=0.05) %>% select(., genes)
myDEindices <- which(genes %in% DEgenes[,1])
exprs <- exprs[,-9]
exprs <- data.matrix(exprs[present,])
hic <- data.matrix(hic[present,])
designy <- as.factor(c("human", "human", "chimp", "chimp", "human", "human", "chimp", "chimp"))
colnames(hic) <- colnames(exprs)
hic <- as.data.table(hic)
exprs <- as.data.table(exprs)
test <- mediate.test.regressing(exprs, designy, hic)
vshrDE <- vash(test$d_se[myDEindices], df=6)
vshrnonDE <- vash(test$d_se[-myDEindices], df=6)
vshrink <- vash(test$d_se, df=6)
DEtest <- test[myDEindices,]
nonDEtest <- test[-myDEindices,]
ash_reg <- ash(as.vector(DEtest$d), as.vector(vshrDE$sd.post), mixcompdist = "normal") #Now it works with this design coding. Use 6 df, as 2-group comparison and 8 individual samples (samples - 2)Due to absence of package REBayes, switching to EM algorithmsummary(ash_reg$result$svalue<=0.05) #Incredible result, looks too good to be true. Mode FALSE TRUE
logical 223 13 ind_sig <- (ash_reg$result$svalue < 0.05)
ind_sig[ind_sig == TRUE] <- "red"
ind_sig[ind_sig == FALSE] <- "black"
plot(x=ash_reg$result$betahat, y=ash_reg$result$sebetahat, ylab="Standard error of the difference", xlab="Difference in effect size from 2 lms, VC", main="Difference of effect size (DE genes)", pch=16, cex=0.6, col=ind_sig)#, xlim=c(-2.5, 2.5), ylim=c(0, 1.3))
legend("topleft", legend=c("s < 0.05", "s >= 0.05"), col=c("red", "black"), pch=16:16, cex=0.8)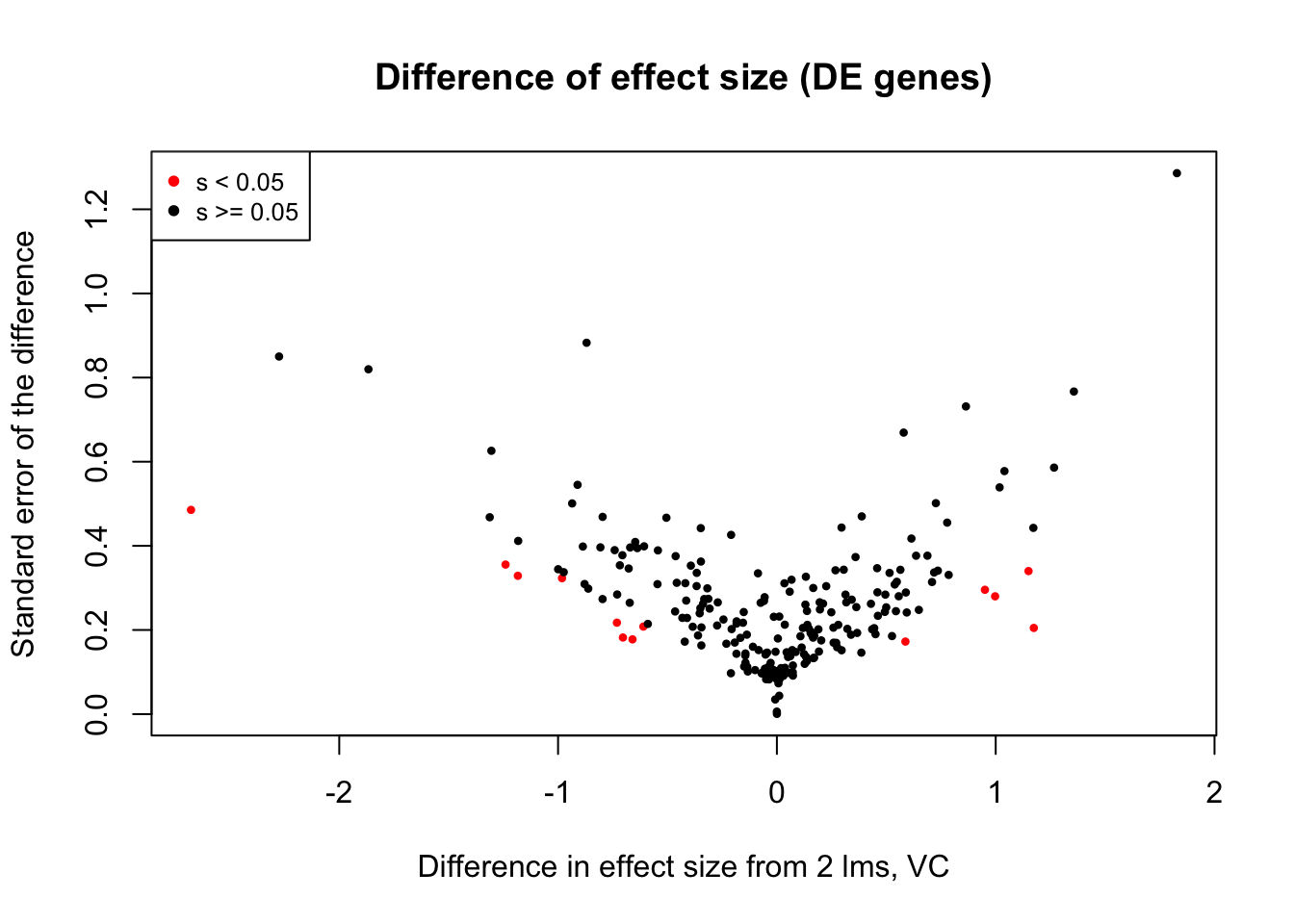
ash_reg_nonDE <- ash(as.vector(nonDEtest$d), as.vector(vshrnonDE$sd.post), mixcompdist = "normal") #Now it works with this design coding. Use 6 df, as 2-group comparison and 8 individual samples (samples - 2)Due to absence of package REBayes, switching to EM algorithmsummary(ash_reg_nonDE$result$svalue<=0.05) #Incredible result, looks too good to be true. Mode FALSE
logical 1394 ind_sig <- (ash_reg_nonDE$result$svalue < 0.05)
ind_sig[ind_sig == TRUE] <- "red"
ind_sig[ind_sig == FALSE] <- "black"
plot(x=ash_reg_nonDE$result$betahat, y=ash_reg_nonDE$result$sebetahat, ylab="Standard error of the difference", xlab="Difference in effect size from 2 lms, VC", main="Difference of effect size (non-DE genes)", pch=16, cex=0.6, col=ind_sig)#, xlim=c(-2.5, 2.5), ylim=c(0, 1.3))
legend("topleft", legend=c("s < 0.05", "s >= 0.05"), col=c("red", "black"), pch=16:16, cex=0.8)
Session information
sessionInfo()R version 3.4.0 (2017-04-21)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: OS X El Capitan 10.11.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] compiler stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] assertthat_0.2.0 bindrcpp_0.2 glmnet_2.0-13
[4] foreach_1.4.4 Matrix_1.2-12 medinome_0.0.1
[7] vashr_0.99.1 qvalue_2.10.0 SQUAREM_2017.10-1
[10] ashr_2.2-7 bedr_1.0.4 forcats_0.3.0
[13] purrr_0.2.4 readr_1.1.1 tibble_1.4.2
[16] tidyverse_1.2.1 edgeR_3.20.9 RColorBrewer_1.1-2
[19] heatmaply_0.14.1 viridis_0.5.0 viridisLite_0.3.0
[22] stringr_1.3.0 gplots_3.0.1 Hmisc_4.1-1
[25] Formula_1.2-2 survival_2.41-3 lattice_0.20-35
[28] dplyr_0.7.4 plotly_4.7.1 cowplot_0.9.3
[31] ggplot2_2.2.1 reshape2_1.4.3 data.table_1.10.4-3
[34] tidyr_0.8.0 plyr_1.8.4 limma_3.34.9
loaded via a namespace (and not attached):
[1] colorspace_1.3-2 class_7.3-14 modeltools_0.2-21
[4] mclust_5.4 rprojroot_1.3-2 htmlTable_1.11.2
[7] futile.logger_1.4.3 base64enc_0.1-3 rstudioapi_0.7
[10] flexmix_2.3-14 mvtnorm_1.0-7 lubridate_1.7.3
[13] xml2_1.2.0 R.methodsS3_1.7.1 codetools_0.2-15
[16] splines_3.4.0 pscl_1.5.2 doParallel_1.0.11
[19] mnormt_1.5-5 robustbase_0.92-8 knitr_1.20
[22] jsonlite_1.5 broom_0.4.3 cluster_2.0.6
[25] kernlab_0.9-25 R.oo_1.21.0 httr_1.3.1
[28] backports_1.1.2 lazyeval_0.2.1 cli_1.0.0
[31] acepack_1.4.1 htmltools_0.3.6 tools_3.4.0
[34] gtable_0.2.0 glue_1.2.0 Rcpp_0.12.16
[37] cellranger_1.1.0 trimcluster_0.1-2 gdata_2.18.0
[40] nlme_3.1-131.1 iterators_1.0.9 fpc_2.1-11
[43] psych_1.7.8 testthat_2.0.0 rvest_0.3.2
[46] gtools_3.5.0 dendextend_1.7.0 DEoptimR_1.0-8
[49] MASS_7.3-49 scales_0.5.0 TSP_1.1-5
[52] hms_0.4.2 parallel_3.4.0 lambda.r_1.2
[55] yaml_2.1.18 gridExtra_2.3 rpart_4.1-13
[58] latticeExtra_0.6-28 stringi_1.1.7 gclus_1.3.1
[61] checkmate_1.8.5 seriation_1.2-3 caTools_1.17.1
[64] truncnorm_1.0-8 rlang_0.2.0 pkgconfig_2.0.1
[67] prabclus_2.2-6 bitops_1.0-6 evaluate_0.10.1
[70] bindr_0.1.1 labeling_0.3 htmlwidgets_1.0
[73] magrittr_1.5 R6_2.2.2 pillar_1.2.1
[76] haven_1.1.1 whisker_0.3-2 foreign_0.8-69
[79] nnet_7.3-12 modelr_0.1.1 crayon_1.3.4
[82] futile.options_1.0.0 KernSmooth_2.23-15 rmarkdown_1.9
[85] locfit_1.5-9.1 grid_3.4.0 readxl_1.0.0
[88] git2r_0.21.0 digest_0.6.15 diptest_0.75-7
[91] webshot_0.5.0 VennDiagram_1.6.19 R.utils_2.6.0
[94] stats4_3.4.0 munsell_0.4.3 registry_0.5 This R Markdown site was created with workflowr